import ROOT
model_ctl =
ROOT.RooAddPdf(
"model_ctl",
"model_ctl", [gx_ctl, px_ctl], [f_ctl])
"combData",
"combined data",
{x},
Index=sample,
Import={"physics": data, "control": data_ctl},
)
simPdf =
ROOT.RooSimultaneous(
"simPdf",
"simultaneous pdf", {
"physics": model,
"control": model_ctl}, sample)
fitResult =
simPdf.fitTo(combData, PrintLevel=-1, Save=
True)
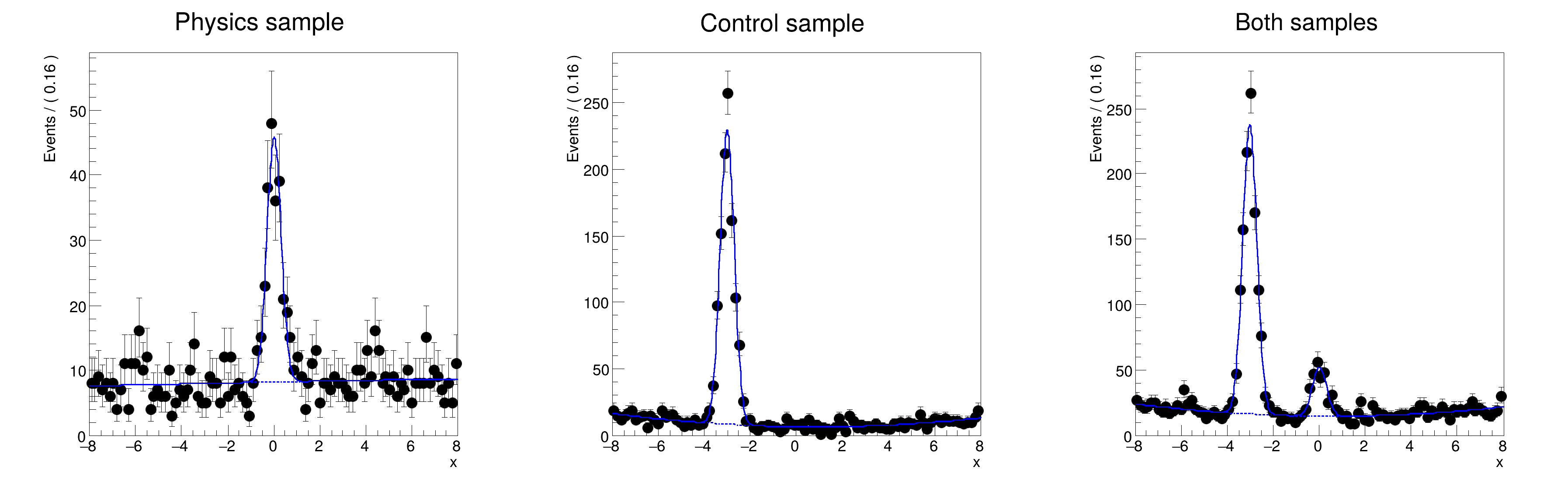
frame1 =
x.frame(Title=
"Physics sample")
simPdf.plotOn(frame1, Slice=(sample,
"physics"), ProjWData=(sample, combData))
simPdf.plotOn(frame1, Slice=(sample,
"physics"), Components=
"px", ProjWData=(sample, combData), LineStyle=
"--")
frame2 =
x.frame(Title=
"Control sample")
simPdf.plotOn(frame2, Components=
"px_ctl", ProjWData=(sample, slicedData), LineStyle=
"--")
frame3 =
x.frame(Title=
"Both samples")
simPdf.plotOn(frame3, Components=
"px,px_ctl", ProjWData=(sample, combData), LineStyle=
"--")
c =
ROOT.TCanvas(
"rf501_simultaneouspdf",
"rf501_simultaneouspdf", 1200, 400)
ROOT::Detail::TRangeCast< T, true > TRangeDynCast
TRangeDynCast is an adapter class that allows the typed iteration through a TCollection.
[#1] INFO:Fitting -- RooAbsPdf::fitTo(simPdf) fixing normalization set for coefficient determination to observables in data
[#1] INFO:Fitting -- using CPU computation library compiled with -mavx512
[#1] INFO:Fitting -- RooAddition::defaultErrorLevel(nll_simPdf_combData) Summation contains a RooNLLVar, using its error level
[#1] INFO:Minimization -- RooAbsMinimizerFcn::setOptimizeConst: activating const optimization
[#1] INFO:Minimization -- RooAbsMinimizerFcn::setOptimizeConst: deactivating const optimization
RooFitResult: minimized FCN value: 8630.62, estimated distance to minimum: 0.000174671
covariance matrix quality: Full, accurate covariance matrix
Status : MINIMIZE=0 HESSE=0
Floating Parameter FinalValue +/- Error
-------------------- --------------------------
a0 6.7634e-02 +/- 6.04e-02
a0_ctl -1.5627e-01 +/- 5.53e-02
a1 -3.8353e-03 +/- 6.32e-02
a1_ctl 3.8442e-01 +/- 4.35e-02
f 1.7952e-01 +/- 1.55e-02
f_ctl 5.2710e-01 +/- 1.25e-02
mean 1.4991e-02 +/- 3.34e-02
mean_ctl -3.0079e+00 +/- 1.04e-02
sigma 3.0450e-01 +/- 8.33e-03
[#1] INFO:Plotting -- RooTreeData::plotOn: plotting 1000 events out of 3000 total events
[#1] INFO:Plotting -- RooSimultaneous::plotOn(simPdf) plot on x represents a slice in the index category (sample)
[#1] INFO:Plotting -- RooAbsReal::plotOn(model) slice variable sample was not projected anyway
[#1] INFO:Plotting -- RooSimultaneous::plotOn(simPdf) plot on x represents a slice in the index category (sample)
[#1] INFO:Plotting -- RooAbsPdf::plotOn(model) directly selected PDF components: (px)
[#1] INFO:Plotting -- RooAbsPdf::plotOn(model) indirectly selected PDF components: ()
[#1] INFO:Plotting -- RooAbsReal::plotOn(model) slice variable sample was not projected anyway
[#1] INFO:Plotting -- RooSimultaneous::plotOn(simPdf) plot on x averages with data index category (sample)
[#1] INFO:Plotting -- RooSimultaneous::plotOn(simPdf) plot on x averages with data index category (sample)
[#1] INFO:Plotting -- RooAbsPdf::plotOn(simPdf) directly selected PDF components: (px_ctl)
[#1] INFO:Plotting -- RooAbsPdf::plotOn(simPdf) indirectly selected PDF components: (model_ctl)
[#1] INFO:Plotting -- RooSimultaneous::plotOn(simPdf) plot on x averages with data index category (sample)
[#1] INFO:Plotting -- RooSimultaneous::plotOn(simPdf) plot on x averages with data index category (sample)
[#1] INFO:Plotting -- RooAbsPdf::plotOn(simPdf) directly selected PDF components: (px,px_ctl)
[#1] INFO:Plotting -- RooAbsPdf::plotOn(simPdf) indirectly selected PDF components: (model_ctl,model)