[#0] WARNING:InputArguments -- The parameter 'sigma1' with range [-inf, inf] of the RooGaussian 'sig1' exceeds the safe range of (0, inf). Advise to limit its range.
[#0] WARNING:InputArguments -- The parameter 'sigma2' with range [-inf, inf] of the RooGaussian 'sig2' exceeds the safe range of (0, inf). Advise to limit its range.
[#0] PROGRESS:Generation -- RooMCStudy::run: sample 990
[#0] PROGRESS:Generation -- RooMCStudy::run: sample 980
[#0] PROGRESS:Generation -- RooMCStudy::run: sample 970
[#0] PROGRESS:Generation -- RooMCStudy::run: sample 960
[#0] PROGRESS:Generation -- RooMCStudy::run: sample 950
RooAbsMinimizerFcn: Minimized function has error status.
Returning maximum FCN so far (-797.721) to force MIGRAD to back out of this region. Error log follows.
Parameter values: a0=0.716236 a1=-0.675686 mean=5.04367 nbkg=115.661 nsig=155.327 sig1frac=0.880121
RooAddPdf::model[ nbkg * bkg_over_bkg_Int[x] + nsig * sig ] has 1 errors
[#0] PROGRESS:Generation -- RooMCStudy::run: sample 940
[#0] PROGRESS:Generation -- RooMCStudy::run: sample 930
RooAbsMinimizerFcn: Minimized function has error status.
Returning maximum FCN so far (-867.289) to force MIGRAD to back out of this region. Error log follows.
Parameter values: a0=0.907715 a1=-0.26012 mean=4.95881 nbkg=147.451 nsig=156.366 sig1frac=0.890535
RooAddPdf::model[ nbkg * bkg_over_bkg_Int[x] + nsig * sig ] has 1 errors
[#0] PROGRESS:Generation -- RooMCStudy::run: sample 920
[#0] PROGRESS:Generation -- RooMCStudy::run: sample 910
[#0] PROGRESS:Generation -- RooMCStudy::run: sample 900
RooAbsMinimizerFcn: Minimized function has error status.
Returning maximum FCN so far (-871.381) to force MIGRAD to back out of this region. Error log follows.
Parameter values: a0=0.716361 a1=-0.355239 mean=5.05578 nbkg=159.647 nsig=151.993 sig1frac=0.742914
RooAddPdf::model[ nbkg * bkg_over_bkg_Int[x] + nsig * sig ] has 1 errors
[#0] PROGRESS:Generation -- RooMCStudy::run: sample 890
[#0] PROGRESS:Generation -- RooMCStudy::run: sample 880
[#0] PROGRESS:Generation -- RooMCStudy::run: sample 870
RooAbsMinimizerFcn: Minimized function has error status.
Returning maximum FCN so far (-903.916) to force MIGRAD to back out of this region. Error log follows.
Parameter values: a0=0.877959 a1=-0.428367 mean=4.99746 nbkg=167.444 nsig=157.288 sig1frac=0.424997
RooAddPdf::model[ nbkg * bkg_over_bkg_Int[x] + nsig * sig ] has 1 errors
[#0] PROGRESS:Generation -- RooMCStudy::run: sample 860
[#0] PROGRESS:Generation -- RooMCStudy::run: sample 850
[#0] PROGRESS:Generation -- RooMCStudy::run: sample 840
[#0] PROGRESS:Generation -- RooMCStudy::run: sample 830
RooAbsMinimizerFcn: Minimized function has error status.
Returning maximum FCN so far (-970.563) to force MIGRAD to back out of this region. Error log follows.
Parameter values: a0=0.867793 a1=-0.296045 mean=5.10353 nbkg=164.822 nsig=173.566 sig1frac=0.899431
RooAddPdf::model[ nbkg * bkg_over_bkg_Int[x] + nsig * sig ] has 1 errors
[#0] PROGRESS:Generation -- RooMCStudy::run: sample 820
[#0] PROGRESS:Generation -- RooMCStudy::run: sample 810
[#0] PROGRESS:Generation -- RooMCStudy::run: sample 800
[#0] PROGRESS:Generation -- RooMCStudy::run: sample 790
[#0] PROGRESS:Generation -- RooMCStudy::run: sample 780
[#0] PROGRESS:Generation -- RooMCStudy::run: sample 770
[#0] PROGRESS:Generation -- RooMCStudy::run: sample 760
RooAbsMinimizerFcn: Minimized function has error status.
Returning maximum FCN so far (-915.472) to force MIGRAD to back out of this region. Error log follows.
Parameter values: a0=0.807508 a1=-0.34963 mean=5.09438 nbkg=142.436 nsig=172.312 sig1frac=0.886183
RooAddPdf::model[ nbkg * bkg_over_bkg_Int[x] + nsig * sig ] has 1 errors
[#0] PROGRESS:Generation -- RooMCStudy::run: sample 750
RooAbsMinimizerFcn: Minimized function has error status.
Returning maximum FCN so far (-885.867) to force MIGRAD to back out of this region. Error log follows.
Parameter values: a0=0.7962 a1=-0.414289 mean=5.07605 nbkg=135.514 nsig=167.164 sig1frac=0.927781
RooAddPdf::model[ nbkg * bkg_over_bkg_Int[x] + nsig * sig ] has 1 errors
RooAbsMinimizerFcn: Minimized function has error status.
Returning maximum FCN so far (-848.597) to force MIGRAD to back out of this region. Error log follows.
Parameter values: a0=0.577138 a1=-0.518554 mean=5.09299 nbkg=144.203 nsig=156.938 sig1frac=0.54541
RooAddPdf::model[ nbkg * bkg_over_bkg_Int[x] + nsig * sig ] has 1 errors
[#0] PROGRESS:Generation -- RooMCStudy::run: sample 740
[#0] PROGRESS:Generation -- RooMCStudy::run: sample 730
[#0] PROGRESS:Generation -- RooMCStudy::run: sample 720
[#0] PROGRESS:Generation -- RooMCStudy::run: sample 710
[#0] PROGRESS:Generation -- RooMCStudy::run: sample 700
RooAbsMinimizerFcn: Minimized function has error status.
Returning maximum FCN so far (-814.549) to force MIGRAD to back out of this region. Error log follows.
Parameter values: a0=0.848973 a1=-0.489149 mean=5.08811 nbkg=132.911 nsig=150.647 sig1frac=0.826731
RooAddPdf::model[ nbkg * bkg_over_bkg_Int[x] + nsig * sig ] has 1 errors
[#0] PROGRESS:Generation -- RooMCStudy::run: sample 690
[#0] PROGRESS:Generation -- RooMCStudy::run: sample 680
[#0] PROGRESS:Generation -- RooMCStudy::run: sample 670
RooAbsMinimizerFcn: Minimized function has error status.
Returning maximum FCN so far (-809.662) to force MIGRAD to back out of this region. Error log follows.
Parameter values: a0=0.713661 a1=-0.606754 mean=5.02862 nbkg=142.583 nsig=145.399 sig1frac=0.623944
RooAddPdf::model[ nbkg * bkg_over_bkg_Int[x] + nsig * sig ] has 1 errors
RooAbsMinimizerFcn: Minimized function has error status.
Returning maximum FCN so far (-861.002) to force MIGRAD to back out of this region. Error log follows.
Parameter values: a0=0.744046 a1=-0.311529 mean=5.09799 nbkg=161.409 nsig=148.884 sig1frac=0.512806
RooAddPdf::model[ nbkg * bkg_over_bkg_Int[x] + nsig * sig ] has 1 errors
[#0] PROGRESS:Generation -- RooMCStudy::run: sample 660
[#0] PROGRESS:Generation -- RooMCStudy::run: sample 650
[#0] PROGRESS:Generation -- RooMCStudy::run: sample 640
[#0] PROGRESS:Generation -- RooMCStudy::run: sample 630
[#0] PROGRESS:Generation -- RooMCStudy::run: sample 620
[#0] PROGRESS:Generation -- RooMCStudy::run: sample 610
RooAbsMinimizerFcn: Minimized function has error status.
Returning maximum FCN so far (-809.027) to force MIGRAD to back out of this region. Error log follows.
Parameter values: a0=0.719517 a1=-0.347533 mean=5.00927 nbkg=127.977 nsig=152.559 sig1frac=0.917291
RooAddPdf::model[ nbkg * bkg_over_bkg_Int[x] + nsig * sig ] has 1 errors
[#0] PROGRESS:Generation -- RooMCStudy::run: sample 600
[#0] PROGRESS:Generation -- RooMCStudy::run: sample 590
[#0] PROGRESS:Generation -- RooMCStudy::run: sample 580
RooAbsMinimizerFcn: Minimized function has error status.
Returning maximum FCN so far (-869.586) to force MIGRAD to back out of this region. Error log follows.
Parameter values: a0=0.621299 a1=-0.442393 mean=5.13262 nbkg=131.7 nsig=167.982 sig1frac=0.776177
RooAddPdf::model[ nbkg * bkg_over_bkg_Int[x] + nsig * sig ] has 1 errors
[#0] PROGRESS:Generation -- RooMCStudy::run: sample 570
[#0] PROGRESS:Generation -- RooMCStudy::run: sample 560
[#0] PROGRESS:Generation -- RooMCStudy::run: sample 550
[#0] PROGRESS:Generation -- RooMCStudy::run: sample 540
[#0] PROGRESS:Generation -- RooMCStudy::run: sample 530
RooAbsMinimizerFcn: Minimized function has error status.
Returning maximum FCN so far (-820.086) to force MIGRAD to back out of this region. Error log follows.
Parameter values: a0=0.699182 a1=-0.532303 mean=4.90748 nbkg=130.722 nsig=155.109 sig1frac=0.843514
RooAddPdf::model[ nbkg * bkg_over_bkg_Int[x] + nsig * sig ] has 1 errors
[#0] PROGRESS:Generation -- RooMCStudy::run: sample 520
[#0] PROGRESS:Generation -- RooMCStudy::run: sample 510
RooAbsMinimizerFcn: Minimized function has error status.
Returning maximum FCN so far (-814.421) to force MIGRAD to back out of this region. Error log follows.
Parameter values: a0=0.729962 a1=-0.49802 mean=4.92039 nbkg=146.276 nsig=144.44 sig1frac=0.620699
RooAddPdf::model[ nbkg * bkg_over_bkg_Int[x] + nsig * sig ] has 1 errors
[#0] PROGRESS:Generation -- RooMCStudy::run: sample 500
[#0] PROGRESS:Generation -- RooMCStudy::run: sample 490
[#0] PROGRESS:Generation -- RooMCStudy::run: sample 480
[#0] PROGRESS:Generation -- RooMCStudy::run: sample 470
[#0] PROGRESS:Generation -- RooMCStudy::run: sample 460
[#0] PROGRESS:Generation -- RooMCStudy::run: sample 450
[#0] PROGRESS:Generation -- RooMCStudy::run: sample 440
[#0] PROGRESS:Generation -- RooMCStudy::run: sample 430
RooAbsMinimizerFcn: Minimized function has error status.
Returning maximum FCN so far (-926.481) to force MIGRAD to back out of this region. Error log follows.
Parameter values: a0=0.653979 a1=-0.469539 mean=5.00078 nbkg=160.963 nsig=166.526 sig1frac=0.628924
RooAddPdf::model[ nbkg * bkg_over_bkg_Int[x] + nsig * sig ] has 1 errors
[#0] PROGRESS:Generation -- RooMCStudy::run: sample 420
RooAbsMinimizerFcn: Minimized function has error status.
Returning maximum FCN so far (-768.847) to force MIGRAD to back out of this region. Error log follows.
Parameter values: a0=0.749295 a1=-0.303934 mean=4.98272 nbkg=121.461 nsig=145.338 sig1frac=0.822833
RooAddPdf::model[ nbkg * bkg_over_bkg_Int[x] + nsig * sig ] has 1 errors
RooAbsMinimizerFcn: Minimized function has error status.
Returning maximum FCN so far (-798.509) to force MIGRAD to back out of this region. Error log follows.
Parameter values: a0=0.791035 a1=-0.287544 mean=4.98693 nbkg=127.577 nsig=149.203 sig1frac=0.914567
RooAddPdf::model[ nbkg * bkg_over_bkg_Int[x] + nsig * sig ] has 1 errors
[#0] PROGRESS:Generation -- RooMCStudy::run: sample 410
[#0] PROGRESS:Generation -- RooMCStudy::run: sample 400
[#0] PROGRESS:Generation -- RooMCStudy::run: sample 390
RooAbsMinimizerFcn: Minimized function has error status.
Returning maximum FCN so far (-977.775) to force MIGRAD to back out of this region. Error log follows.
Parameter values: a0=0.565967 a1=-0.529635 mean=5.0065 nbkg=153.38 nsig=185.083 sig1frac=0.751239
RooAddPdf::model[ nbkg * bkg_over_bkg_Int[x] + nsig * sig ] has 1 errors
[#0] PROGRESS:Generation -- RooMCStudy::run: sample 380
[#0] PROGRESS:Generation -- RooMCStudy::run: sample 370
RooAbsMinimizerFcn: Minimized function has error status.
Returning maximum FCN so far (-945.036) to force MIGRAD to back out of this region. Error log follows.
Parameter values: a0=0.824344 a1=-0.338546 mean=5.06339 nbkg=161.705 nsig=169.087 sig1frac=0.910016
RooAddPdf::model[ nbkg * bkg_over_bkg_Int[x] + nsig * sig ] has 1 errors
[#0] PROGRESS:Generation -- RooMCStudy::run: sample 360
[#0] PROGRESS:Generation -- RooMCStudy::run: sample 350
[#0] PROGRESS:Generation -- RooMCStudy::run: sample 340
[#0] PROGRESS:Generation -- RooMCStudy::run: sample 330
[#0] PROGRESS:Generation -- RooMCStudy::run: sample 320
[#0] PROGRESS:Generation -- RooMCStudy::run: sample 310
RooAbsMinimizerFcn: Minimized function has error status.
Returning maximum FCN so far (-831.776) to force MIGRAD to back out of this region. Error log follows.
Parameter values: a0=0.823781 a1=-0.287947 mean=4.96347 nbkg=133.185 nsig=155.535 sig1frac=0.887106
RooAddPdf::model[ nbkg * bkg_over_bkg_Int[x] + nsig * sig ] has 1 errors
[#0] PROGRESS:Generation -- RooMCStudy::run: sample 300
[#0] PROGRESS:Generation -- RooMCStudy::run: sample 290
[#0] PROGRESS:Generation -- RooMCStudy::run: sample 280
RooAbsMinimizerFcn: Minimized function has error status.
Returning maximum FCN so far (-867.974) to force MIGRAD to back out of this region. Error log follows.
Parameter values: a0=0.806741 a1=-0.376486 mean=4.95962 nbkg=143.191 nsig=160.421 sig1frac=0.787531
RooAddPdf::model[ nbkg * bkg_over_bkg_Int[x] + nsig * sig ] has 1 errors
[#0] PROGRESS:Generation -- RooMCStudy::run: sample 270
RooAbsMinimizerFcn: Minimized function has error status.
Returning maximum FCN so far (-810.184) to force MIGRAD to back out of this region. Error log follows.
Parameter values: a0=0.701119 a1=-0.376392 mean=4.96099 nbkg=126.85 nsig=153.527 sig1frac=0.880449
RooAddPdf::model[ nbkg * bkg_over_bkg_Int[x] + nsig * sig ] has 1 errors
RooAbsMinimizerFcn: Minimized function has error status.
Returning maximum FCN so far (-845.198) to force MIGRAD to back out of this region. Error log follows.
Parameter values: a0=0.74363 a1=-0.435344 mean=5.01952 nbkg=162.467 nsig=143.571 sig1frac=0.630815
RooAddPdf::model[ nbkg * bkg_over_bkg_Int[x] + nsig * sig ] has 1 errors
[#0] PROGRESS:Generation -- RooMCStudy::run: sample 260
[#0] PROGRESS:Generation -- RooMCStudy::run: sample 250
RooAbsMinimizerFcn: Minimized function has error status.
Returning maximum FCN so far (-1003.82) to force MIGRAD to back out of this region. Error log follows.
Parameter values: a0=0.674666 a1=-0.383697 mean=5.00365 nbkg=169.829 nsig=179.534 sig1frac=0.921743
RooAddPdf::model[ nbkg * bkg_over_bkg_Int[x] + nsig * sig ] has 1 errors
RooAbsMinimizerFcn: Minimized function has error status.
Returning maximum FCN so far (-712.949) to force MIGRAD to back out of this region. Error log follows.
Parameter values: a0=0.711808 a1=-0.34088 mean=4.99573 nbkg=115.49 nsig=133.015 sig1frac=0.908627
RooAddPdf::model[ nbkg * bkg_over_bkg_Int[x] + nsig * sig ] has 1 errors
[#0] PROGRESS:Generation -- RooMCStudy::run: sample 240
[#0] PROGRESS:Generation -- RooMCStudy::run: sample 230
[#0] PROGRESS:Generation -- RooMCStudy::run: sample 220
[#0] PROGRESS:Generation -- RooMCStudy::run: sample 210
[#0] PROGRESS:Generation -- RooMCStudy::run: sample 200
[#0] PROGRESS:Generation -- RooMCStudy::run: sample 190
[#0] PROGRESS:Generation -- RooMCStudy::run: sample 180
[#0] PROGRESS:Generation -- RooMCStudy::run: sample 170
[#0] PROGRESS:Generation -- RooMCStudy::run: sample 160
[#0] PROGRESS:Generation -- RooMCStudy::run: sample 150
RooAbsMinimizerFcn: Minimized function has error status.
Returning maximum FCN so far (-750.903) to force MIGRAD to back out of this region. Error log follows.
Parameter values: a0=0.726225 a1=-0.429497 mean=5.03871 nbkg=129.53 nsig=136.644 sig1frac=0.687558
RooAddPdf::model[ nbkg * bkg_over_bkg_Int[x] + nsig * sig ] has 1 errors
[#0] PROGRESS:Generation -- RooMCStudy::run: sample 140
RooAbsMinimizerFcn: Minimized function has error status.
Returning maximum FCN so far (-740.756) to force MIGRAD to back out of this region. Error log follows.
Parameter values: a0=0.899536 a1=-0.345881 mean=5.03897 nbkg=135.176 nsig=129.13 sig1frac=0.899393
RooAddPdf::model[ nbkg * bkg_over_bkg_Int[x] + nsig * sig ] has 1 errors
RooAbsMinimizerFcn: Minimized function has error status.
Returning maximum FCN so far (-677.604) to force MIGRAD to back out of this region. Error log follows.
Parameter values: a0=0.766597 a1=-0.547838 mean=5.03836 nbkg=119.039 nsig=121.817 sig1frac=0.213703
RooAddPdf::model[ nbkg * bkg_over_bkg_Int[x] + nsig * sig ] has 1 errors
[#0] PROGRESS:Generation -- RooMCStudy::run: sample 130
RooAbsMinimizerFcn: Minimized function has error status.
Returning maximum FCN so far (-777.498) to force MIGRAD to back out of this region. Error log follows.
Parameter values: a0=0.527582 a1=-0.893856 mean=5.24753 nbkg=162.951 nsig=65.9357 sig1frac=0.970262
RooAddPdf::model[ nbkg * bkg_over_bkg_Int[x] + nsig * sig ] has 1 errors
RooAbsMinimizerFcn: Minimized function has error status.
Returning maximum FCN so far (-777.498) to force MIGRAD to back out of this region. Error log follows.
Parameter values: a0=0.516095 a1=-0.955515 mean=5.14443 nbkg=157.502 nsig=97.2821 sig1frac=0.174928
RooAddPdf::model[ nbkg * bkg_over_bkg_Int[x] + nsig * sig ] has 1 errors
RooAbsMinimizerFcn: Minimized function has error status.
Returning maximum FCN so far (-777.498) to force MIGRAD to back out of this region. Error log follows.
Parameter values: a0=0.510348 a1=-0.776595 mean=5.09286 nbkg=154.805 nsig=114.979 sig1frac=0.0496841
RooAddPdf::model[ nbkg * bkg_over_bkg_Int[x] + nsig * sig ] has 1 errors
[#0] PROGRESS:Generation -- RooMCStudy::run: sample 120
[#0] PROGRESS:Generation -- RooMCStudy::run: sample 110
RooAbsMinimizerFcn: Minimized function has error status.
Returning maximum FCN so far (-796.54) to force MIGRAD to back out of this region. Error log follows.
Parameter values: a0=0.702991 a1=-0.414346 mean=5.11911 nbkg=145.466 nsig=141.134 sig1frac=0.532895
RooAddPdf::model[ nbkg * bkg_over_bkg_Int[x] + nsig * sig ] has 1 errors
[#0] PROGRESS:Generation -- RooMCStudy::run: sample 100
RooAbsMinimizerFcn: Minimized function has error status.
Returning maximum FCN so far (-920.777) to force MIGRAD to back out of this region. Error log follows.
Parameter values: a0=0.726241 a1=-0.424659 mean=5.05565 nbkg=151.228 nsig=170.816 sig1frac=0.771802
RooAddPdf::model[ nbkg * bkg_over_bkg_Int[x] + nsig * sig ] has 1 errors
RooAbsMinimizerFcn: Minimized function has error status.
Returning maximum FCN so far (-743.339) to force MIGRAD to back out of this region. Error log follows.
Parameter values: a0=0.942558 a1=-0.775768 mean=4.99866 nbkg=187.872 nsig=63.0852 sig1frac=0.868175
RooAddPdf::model[ nbkg * bkg_over_bkg_Int[x] + nsig * sig ] has 1 errors
RooAbsMinimizerFcn: Minimized function has error status.
Returning maximum FCN so far (-743.339) to force MIGRAD to back out of this region. Error log follows.
Parameter values: a0=0.984478 a1=-0.59938 mean=4.99914 nbkg=173.959 nsig=90.3182 sig1frac=0.461117
RooAddPdf::model[ nbkg * bkg_over_bkg_Int[x] + nsig * sig ] has 1 errors
[#0] PROGRESS:Generation -- RooMCStudy::run: sample 90
[#0] PROGRESS:Generation -- RooMCStudy::run: sample 80
[#0] PROGRESS:Generation -- RooMCStudy::run: sample 70
RooAbsMinimizerFcn: Minimized function has error status.
Returning maximum FCN so far (-775.389) to force MIGRAD to back out of this region. Error log follows.
Parameter values: a0=0.72761 a1=-0.375581 mean=5.04879 nbkg=141.143 nsig=137.402 sig1frac=0.495257
RooAddPdf::model[ nbkg * bkg_over_bkg_Int[x] + nsig * sig ] has 1 errors
[#0] PROGRESS:Generation -- RooMCStudy::run: sample 60
[#0] PROGRESS:Generation -- RooMCStudy::run: sample 50
[#0] PROGRESS:Generation -- RooMCStudy::run: sample 40
[#0] PROGRESS:Generation -- RooMCStudy::run: sample 30
[#0] PROGRESS:Generation -- RooMCStudy::run: sample 20
[#0] PROGRESS:Generation -- RooMCStudy::run: sample 10
[#0] PROGRESS:Generation -- RooMCStudy::run: sample 0
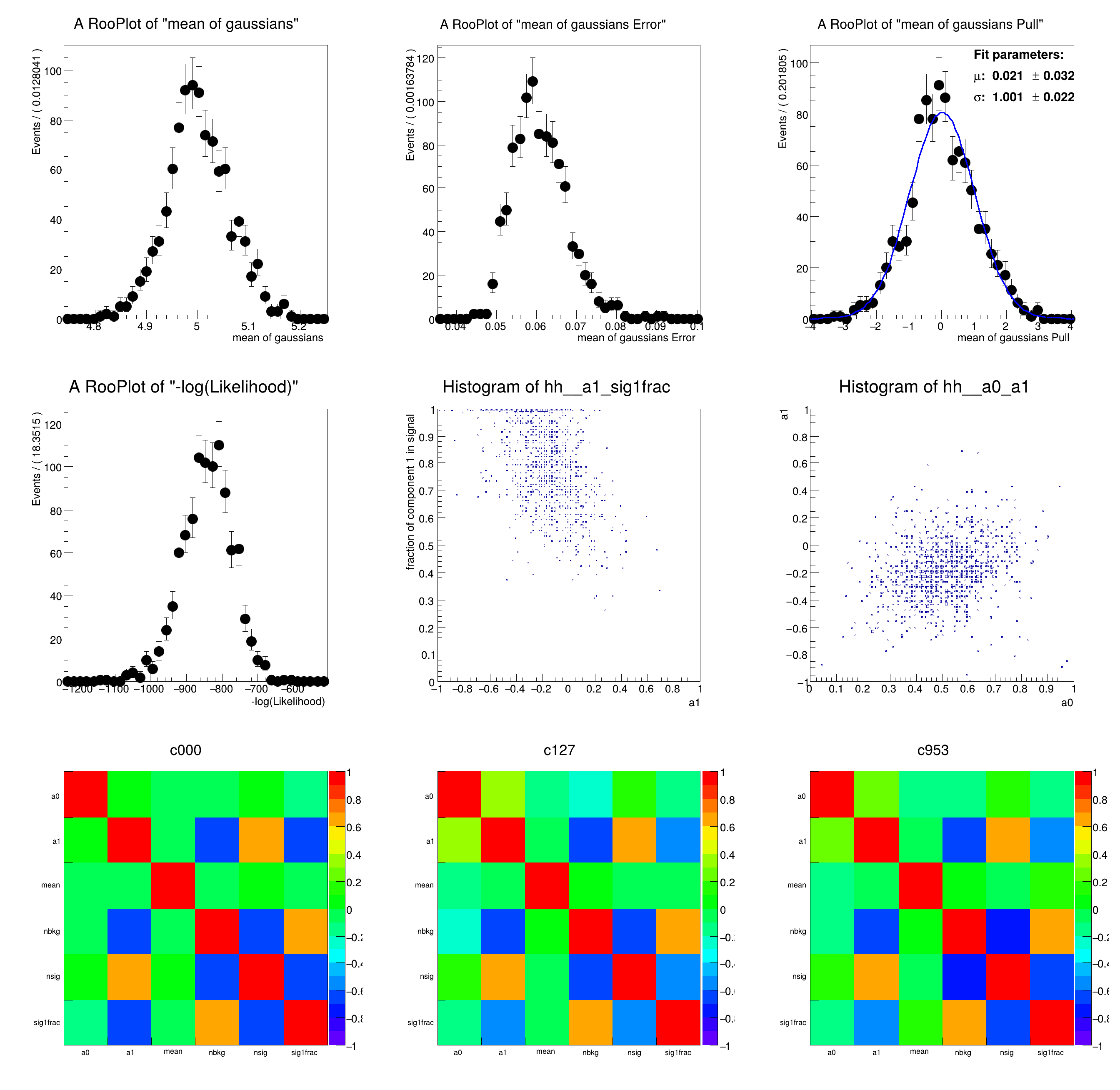
[#0] WARNING:Generation -- Fit parameter 'sigma1' does not have an error. A pull distribution cannot be generated. This might be caused by the parameter being constant or because the fits were not run.
[#0] WARNING:Generation -- Fit parameter 'sigma2' does not have an error. A pull distribution cannot be generated. This might be caused by the parameter being constant or because the fits were not run.
[#1] INFO:ObjectHandling -- RooWorkspace::import() importing RooRealVar::meanpull
[#1] INFO:Fitting -- RooAbsPdf::fitTo(pullGauss_over_pullGauss_Int[meanpull]) fixing normalization set for coefficient determination to observables in data
[#1] INFO:Fitting -- using CPU computation library compiled with -mavx512
[#1] INFO:Fitting -- RooAddition::defaultErrorLevel(nll_pullGauss_over_pullGauss_Int[meanpull]_fitParData_model) Summation contains a RooNLLVar, using its error level
[#1] INFO:Minimization -- RooAbsMinimizerFcn::setOptimizeConst: activating const optimization
[#1] INFO:Minimization -- RooAbsMinimizerFcn::setOptimizeConst: deactivating const optimization