RooWorkspace(combined) combined contents
variables
---------
(Lumi,SigXsecOverSM,alpha_syst1,alpha_syst2,alpha_syst3,channelCat,gamma_stat_channel1_bin_0,gamma_stat_channel1_bin_1,nom_alpha_syst1,nom_alpha_syst2,nom_alpha_syst3,nom_gamma_stat_channel1_bin_0,nom_gamma_stat_channel1_bin_1,nominalLumi,obs_x_channel1)
p.d.f.s
-------
RooGaussian::alpha_syst1Constraint[ x=alpha_syst1 mean=nom_alpha_syst1 sigma=1 ] = 1
RooGaussian::alpha_syst2Constraint[ x=alpha_syst2 mean=nom_alpha_syst2 sigma=1 ] = 1
RooGaussian::alpha_syst3Constraint[ x=alpha_syst3 mean=nom_alpha_syst3 sigma=1 ] = 1
RooRealSumPdf::channel1_model[ signal_channel1_scaleFactors * signal_channel1_shapes + background1_channel1_scaleFactors * background1_channel1_shapes + background2_channel1_scaleFactors * background2_channel1_shapes ] = 240
RooPoisson::gamma_stat_channel1_bin_0_constraint[ x=nom_gamma_stat_channel1_bin_0 mean=gamma_stat_channel1_bin_0_poisMean ] = 0.019943
RooPoisson::gamma_stat_channel1_bin_1_constraint[ x=nom_gamma_stat_channel1_bin_1 mean=gamma_stat_channel1_bin_1_poisMean ] = 0.039861
RooGaussian::lumiConstraint[ x=Lumi mean=nominalLumi sigma=0.1 ] = 1
RooProdPdf::model_channel1[ lumiConstraint * alpha_syst1Constraint * alpha_syst2Constraint * alpha_syst3Constraint * gamma_stat_channel1_bin_0_constraint * gamma_stat_channel1_bin_1_constraint * channel1_model(obs_x_channel1) ] = 0.190787
RooSimultaneous::simPdf[ indexCat=channelCat channel1=model_channel1 ] = 0.190787
functions
--------
RooHistFunc::background1_channel1_Hist_alphanominal[ depList=(obs_x_channel1) depList=(obs_x_channel1) ] = 100
RooStats::HistFactory::FlexibleInterpVar::background1_channel1_epsilon[ paramList=(alpha_syst2) ] = 1
RooProduct::background1_channel1_scaleFactors[ background1_channel1_epsilon * Lumi ] = 1
RooProduct::background1_channel1_shapes[ background1_channel1_Hist_alphanominal * mc_stat_channel1 * channel1_model_binWidth ] = 200
RooHistFunc::background2_channel1_Hist_alphanominal[ depList=(obs_x_channel1) depList=(obs_x_channel1) ] = 0
RooStats::HistFactory::FlexibleInterpVar::background2_channel1_epsilon[ paramList=(alpha_syst3) ] = 1
RooProduct::background2_channel1_scaleFactors[ background2_channel1_epsilon * Lumi ] = 1
RooProduct::background2_channel1_shapes[ background2_channel1_Hist_alphanominal * mc_stat_channel1 * channel1_model_binWidth ] = 0
RooBinWidthFunction::channel1_model_binWidth[ HistFuncForBinWidth=signal_channel1_Hist_alphanominal HistFuncForBinWidth=signal_channel1_Hist_alphanominal ] = 2
RooProduct::gamma_stat_channel1_bin_0_poisMean[ gamma_stat_channel1_bin_0 * gamma_stat_channel1_bin_0_tau ] = 400
RooProduct::gamma_stat_channel1_bin_1_poisMean[ gamma_stat_channel1_bin_1 * gamma_stat_channel1_bin_1_tau ] = 100
ParamHistFunc::mc_stat_channel1[ ] = 1
RooHistFunc::signal_channel1_Hist_alphanominal[ depList=(obs_x_channel1) depList=(obs_x_channel1) ] = 20
RooStats::HistFactory::FlexibleInterpVar::signal_channel1_epsilon[ paramList=(alpha_syst1) ] = 1
RooProduct::signal_channel1_scaleFactors[ signal_channel1_epsilon * SigXsecOverSM * Lumi ] = 1
RooProduct::signal_channel1_shapes[ signal_channel1_Hist_alphanominal * channel1_model_binWidth ] = 40
datasets
--------
RooDataSet::obsData(obs_x_channel1,channelCat)
RooDataSet::asimovData(obs_x_channel1,channelCat)
embedded datasets (in pdfs and functions)
-----------------------------------------
RooDataHist::signal_channel1_Hist_alphanominalDHist(obs_x_channel1)
RooDataHist::background1_channel1_Hist_alphanominalDHist(obs_x_channel1)
RooDataHist::background2_channel1_Hist_alphanominalDHist(obs_x_channel1)
parameter snapshots
-------------------
NominalParamValues = (nominalLumi=1[C],nom_alpha_syst1=0[C],nom_alpha_syst2=0[C],nom_alpha_syst3=0[C],nom_gamma_stat_channel1_bin_0=400[C],nom_gamma_stat_channel1_bin_1=100[C],obs_x_channel1=1.25,Lumi=1 +/- 0.1[C],alpha_syst1=0 +/- 1[C],alpha_syst2=0 +/- 1,alpha_syst3=0 +/- 1,gamma_stat_channel1_bin_0=1 +/- 0.05,gamma_stat_channel1_bin_1=1 +/- 0.1,SigXsecOverSM=1 +/- 0)
named sets
----------
ModelConfig_GlobalObservables:(nominalLumi,nom_alpha_syst1,nom_alpha_syst2,nom_alpha_syst3,nom_gamma_stat_channel1_bin_0,nom_gamma_stat_channel1_bin_1)
ModelConfig_NuisParams:(alpha_syst2,alpha_syst3,gamma_stat_channel1_bin_0,gamma_stat_channel1_bin_1)
ModelConfig_Observables:(obs_x_channel1,channelCat)
ModelConfig_POI:(SigXsecOverSM)
globalObservables:(nominalLumi,nom_alpha_syst1,nom_alpha_syst2,nom_alpha_syst3,nom_gamma_stat_channel1_bin_0,nom_gamma_stat_channel1_bin_1)
observables:(obs_x_channel1,channelCat)
generic objects
---------------
RooStats::ModelConfig::ModelConfig
=== Using the following for ModelConfigB_only ===
Observables: RooArgSet:: = (obs_x_channel1,channelCat)
Parameters of Interest: RooArgSet:: = (SigXsecOverSM)
Nuisance Parameters: RooArgSet:: = (alpha_syst2,alpha_syst3,gamma_stat_channel1_bin_0,gamma_stat_channel1_bin_1)
Global Observables: RooArgSet:: = (nominalLumi,nom_alpha_syst1,nom_alpha_syst2,nom_alpha_syst3,nom_gamma_stat_channel1_bin_0,nom_gamma_stat_channel1_bin_1)
PDF: RooSimultaneous::simPdf[ indexCat=channelCat channel1=model_channel1 ] = 0.190787
Snapshot:
1) 0x77d5ae0 RooRealVar:: SigXsecOverSM = 0 +/- 0 L(0 - 3) "SigXsecOverSM"
=== Using the following for ModelConfig ===
Observables: RooArgSet:: = (obs_x_channel1,channelCat)
Parameters of Interest: RooArgSet:: = (SigXsecOverSM)
Nuisance Parameters: RooArgSet:: = (alpha_syst2,alpha_syst3,gamma_stat_channel1_bin_0,gamma_stat_channel1_bin_1)
Global Observables: RooArgSet:: = (nominalLumi,nom_alpha_syst1,nom_alpha_syst2,nom_alpha_syst3,nom_gamma_stat_channel1_bin_0,nom_gamma_stat_channel1_bin_1)
PDF: RooSimultaneous::simPdf[ indexCat=channelCat channel1=model_channel1 ] = 0.190787
Snapshot:
1) 0x77d60f0 RooRealVar:: SigXsecOverSM = 1 +/- 0 L(0 - 3) "SigXsecOverSM"
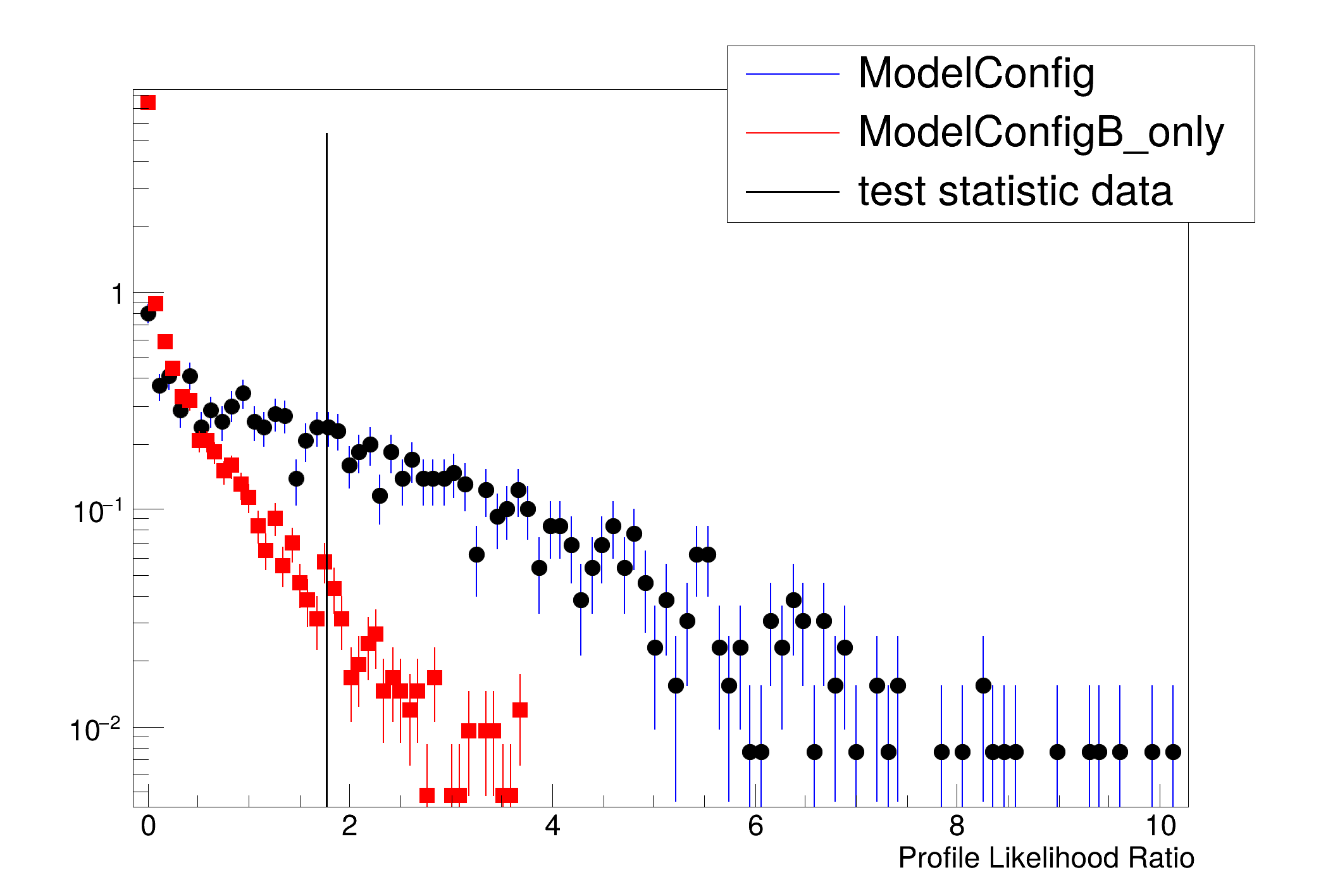
[#0] PROGRESS:Generation -- Test Statistic on data: 1.77388
[#1] INFO:InputArguments -- Profiling conditional MLEs for Null.
[#1] INFO:InputArguments -- Using a ToyMCSampler. Now configuring for Null.
[#0] PROGRESS:Generation -- generated toys: 500 / 5000
[#0] PROGRESS:Generation -- generated toys: 1000 / 5000
[#0] PROGRESS:Generation -- generated toys: 1500 / 5000
[#0] PROGRESS:Generation -- generated toys: 2000 / 5000
[#0] PROGRESS:Generation -- generated toys: 2500 / 5000
[#0] PROGRESS:Generation -- generated toys: 3000 / 5000
[#0] PROGRESS:Generation -- generated toys: 3500 / 5000
[#0] PROGRESS:Generation -- generated toys: 4000 / 5000
[#0] PROGRESS:Generation -- generated toys: 4500 / 5000
[#1] INFO:InputArguments -- Profiling conditional MLEs for Alt.
[#1] INFO:InputArguments -- Using a ToyMCSampler. Now configuring for Alt.
[#0] PROGRESS:Generation -- generated toys: 500 / 1250
[#0] PROGRESS:Generation -- generated toys: 1000 / 1250
Results HypoTestCalculator_result:
- Null p-value = 0.0306 +/- 0.00243572
- Significance = 1.87205 +/- 0.0352146 sigma
- Number of Alt toys: 1250
- Number of Null toys: 5000
- Test statistic evaluated on data: 1.77388
- CL_b: 0.0306 +/- 0.00243572
- CL_s+b: 0.4312 +/- 0.0140076
- CL_s: 14.0915 +/- 1.21148
Expected p -value and significance at -2 sigma = 0.7916 significance -0.811985 sigma
Expected p -value and significance at -1 sigma = 0.2472 significance 0.683327 sigma
Expected p -value and significance at 0 sigma = 0.0434 significance 1.71252 sigma
Expected p -value and significance at 1 sigma = 0.0034 significance 2.70648 sigma
Expected p -value and significance at 2 sigma = 0.0004 significance 3.35279 sigma
#include <cassert>
bool generateBinned = false;
};
{
bool generateBinned =
optHT.generateBinned;
SimpleLikelihoodRatioTestStat::SetAlwaysReuseNLL(true);
ProfileLikelihoodTestStat::SetAlwaysReuseNLL(true);
RatioOfProfiledLikelihoodsTestStat::SetAlwaysReuseNLL(true);
filename =
"results/example_combined_GaussExample_model.root";
cout << "will run standard hist2workspace example" << endl;
gROOT->ProcessLine(
".! prepareHistFactory .");
gROOT->ProcessLine(
".! hist2workspace config/example.xml");
cout << "\n\n---------------------" << endl;
cout << "Done creating example input" << endl;
cout << "---------------------\n\n" << endl;
}
} else
if (!file) {
cout <<
"StandardRooStatsDemoMacro: Input file " <<
filename <<
" is not found" << endl;
return;
}
cout << "workspace not found" << endl;
return;
}
cout << "data or ModelConfig was not found" << endl;
return;
}
std::cout << "StandardHypoTestInvDemo"
<< " - Switch off all systematics by setting them constant to their initial values" << std::endl;
}
}
}
Info(
"StandardHypoTestInvDemo",
"The background model %s does not exist",
modelBName);
Info(
"StandardHypoTestInvDemo",
"Copy it from ModelConfig %s and set POI to zero",
modelSBName);
if (!var)
return;
}
Info(
"StandardHypoTestDemo",
"Model %s has no snapshot - make one using model poi",
modelSBName);
if (!var)
return;
}
if (
bModel->GetNuisanceParameters())
if (
sbModel->GetNuisanceParameters())
ropl->SetSubtractMLE(
false);
profll->SetOneSidedDiscovery(1);
slrts->EnableDetailedOutput();
profll->EnableDetailedOutput();
ropl->EnableDetailedOutput();
}
}
}
"Only the PL test statistic can be used with AsymptoticCalculator - use by default a two-sided PL");
}
Info(
"StandardHypoTestDemo",
"No nuisance pdf given for the HybridCalculator - try to deduce pdf from the model");
else
}
Info(
"StandardHypoTestDemo",
"No nuisance pdf given - try to use %s that is defined as a prior pdf in the B model",
} else {
Error(
"StandardHypoTestDemo",
"Cannot run Hybrid calculator because no prior on the nuisance parameter is "
"specified or can be derived");
return;
}
}
Info(
"StandardHypoTestDemo",
"Using as nuisance Pdf ... ");
(
bModel->GetNuisanceParameters()) ?
bModel->GetNuisanceParameters() :
sbModel->GetNuisanceParameters();
if (
np->getSize() == 0) {
"Prior nuisance does not depend on nuisance parameters. They will be smeared in their full range");
}
}
if (
sbModel->GetPdf()->canBeExtended()) {
Warning(
"StandardHypoTestDemo",
"Pdf is extended: but number counting flag is set: ignore it ");
} else {
int nEvents =
data->numEntries();
Info(
"StandardHypoTestDemo",
"Pdf is not extended: number of events to generate taken from observed data set is %d", nEvents);
sampler->SetNEventsPerToy(nEvents);
} else {
Info(
"StandardHypoTestDemo",
"using a number counting pdf");
}
}
if (
data->isWeighted() && !generateBinned) {
Info(
"StandardHypoTestDemo",
"Data set is weighted, nentries = %d and sum of weights = %8.1f but toy "
"generation is unbinned - it would be faster to set generateBinned to true\n",
data->numEntries(),
data->sumEntries());
}
if (generateBinned)
sampler->SetGenerateBinned(generateBinned);
}
htr->SetPValueIsRightTail(
true);
htr->SetBackgroundAsAlt(
false);
} else {
std::cout << "Asymptotic results " << std::endl;
}
for (int i = 0; i < 5; ++i) {
double sig = -2 + i;
}
std::vector<double> values =
altDist->GetSamplingDistribution();
for (int i = 0; i < 5; ++i) {
htExp.SetTestStatisticData(
q[i]);
double sig = -2 + i;
std::cout <<
" Expected p -value and significance at " << sig <<
" sigma = " <<
htExp.NullPValue()
<<
" significance " <<
htExp.Significance() <<
" sigma " << std::endl;
}
} else {
for (int i = 0; i < 5; ++i) {
double sig = -2 + i;
double pval = AsymptoticCalculator::GetExpectedPValues(
htr->NullPValue(),
htr->AlternatePValue(), -sig,
false);
std::cout <<
" Expected p -value and significance at " << sig <<
" sigma = " <<
pval <<
" significance " }
}
Info(
"StandardHypoTestDemo",
"Detailed output will be written in output result file");
}
name.Replace(0,
name.Last(
'/') + 1,
"");
Info(
"StandardHypoTestDemo",
"HypoTestResult has been written in the file %s",
resultFileName.Data());
fileOut->Close();
}
}
void Info(const char *location, const char *msgfmt,...)
Use this function for informational messages.
void Error(const char *location, const char *msgfmt,...)
Use this function in case an error occurred.
void Warning(const char *location, const char *msgfmt,...)
Use this function in warning situations.
winID h TVirtualViewer3D TVirtualGLPainter p
winID h TVirtualViewer3D TVirtualGLPainter char TVirtualGLPainter plot
Option_t Option_t TPoint TPoint const char GetTextMagnitude GetFillStyle GetLineColor GetLineWidth GetMarkerStyle GetTextAlign GetTextColor GetTextSize void char Point_t Rectangle_t WindowAttributes_t Float_t Float_t Float_t Int_t Int_t UInt_t UInt_t Rectangle_t Int_t Int_t Window_t TString Int_t GCValues_t GetPrimarySelectionOwner GetDisplay GetScreen GetColormap GetNativeEvent const char const char dpyName wid window const char font_name cursor keysym reg const char only_if_exist regb h Point_t winding char text const char depth char const char Int_t count const char ColorStruct_t color const char filename
Option_t Option_t TPoint TPoint const char GetTextMagnitude GetFillStyle GetLineColor GetLineWidth GetMarkerStyle GetTextAlign GetTextColor GetTextSize void char Point_t Rectangle_t WindowAttributes_t Float_t Float_t Float_t Int_t Int_t UInt_t UInt_t Rectangle_t Int_t Int_t Window_t TString Int_t GCValues_t GetPrimarySelectionOwner GetDisplay GetScreen GetColormap GetNativeEvent const char const char dpyName wid window const char font_name cursor keysym reg const char only_if_exist regb h Point_t np
R__EXTERN TSystem * gSystem
Abstract base class for binned and unbinned datasets.
Abstract interface for all probability density functions.
double getVal(const RooArgSet *normalisationSet=nullptr) const
Evaluate object.
RooArgSet is a container object that can hold multiple RooAbsArg objects.
Variable that can be changed from the outside.
void setVal(double value) override
Set value of variable to 'value'.
Hypothesis Test Calculator based on the asymptotic formulae for the profile likelihood ratio.
Does a frequentist hypothesis test.
Same purpose as HybridCalculatorOriginal, but different implementation.
Common base class for the Hypothesis Test Calculators.
This class provides the plots for the result of a study performed with any of the HypoTestCalculatorG...
HypoTestResult is a base class for results from hypothesis tests.
< A class that holds configuration information for a model using a workspace as a store
ProfileLikelihoodTestStat is an implementation of the TestStatistic interface that calculates the pro...
TestStatistic that returns the ratio of profiled likelihoods.
This class simply holds a sampling distribution of some test statistic.
TestStatistic class that returns -log(L[null] / L[alt]) where L is the likelihood.
ToyMCSampler is an implementation of the TestStatSampler interface.
Persistable container for RooFit projects.
TObject * Get(const char *namecycle) override
Return pointer to object identified by namecycle.
A ROOT file is an on-disk file, usually with extension .root, that stores objects in a file-system-li...
static TFile * Open(const char *name, Option_t *option="", const char *ftitle="", Int_t compress=ROOT::RCompressionSetting::EDefaults::kUseCompiledDefault, Int_t netopt=0)
Create / open a file.
static TString Format(const char *fmt,...)
Static method which formats a string using a printf style format descriptor and return a TString.
virtual Bool_t AccessPathName(const char *path, EAccessMode mode=kFileExists)
Returns FALSE if one can access a file using the specified access mode.
double normal_cdf(double x, double sigma=1, double x0=0)
Cumulative distribution function of the normal (Gaussian) distribution (lower tail).
double normal_quantile_c(double z, double sigma)
Inverse ( ) of the cumulative distribution function of the upper tail of the normal (Gaussian) distri...
The namespace RooFit contains mostly switches that change the behaviour of functions of PDFs (or othe...
Namespace for the RooStats classes.
bool SetAllConstant(const RooAbsCollection &coll, bool constant=true)
utility function to set all variable constant in a collection (from G.
RooAbsPdf * MakeNuisancePdf(RooAbsPdf &pdf, const RooArgSet &observables, const char *name)
extract constraint terms from pdf
void Quantiles(Int_t n, Int_t nprob, Double_t *x, Double_t *quantiles, Double_t *prob, Bool_t isSorted=kTRUE, Int_t *index=nullptr, Int_t type=7)
Computes sample quantiles, corresponding to the given probabilities.