This tutorial produces an N-dimensional multivariate Gaussian with a non-trivial covariance matrix. By default N=4 (called "dim").
A subset of these are considered parameters of interest. This problem is tractable analytically.
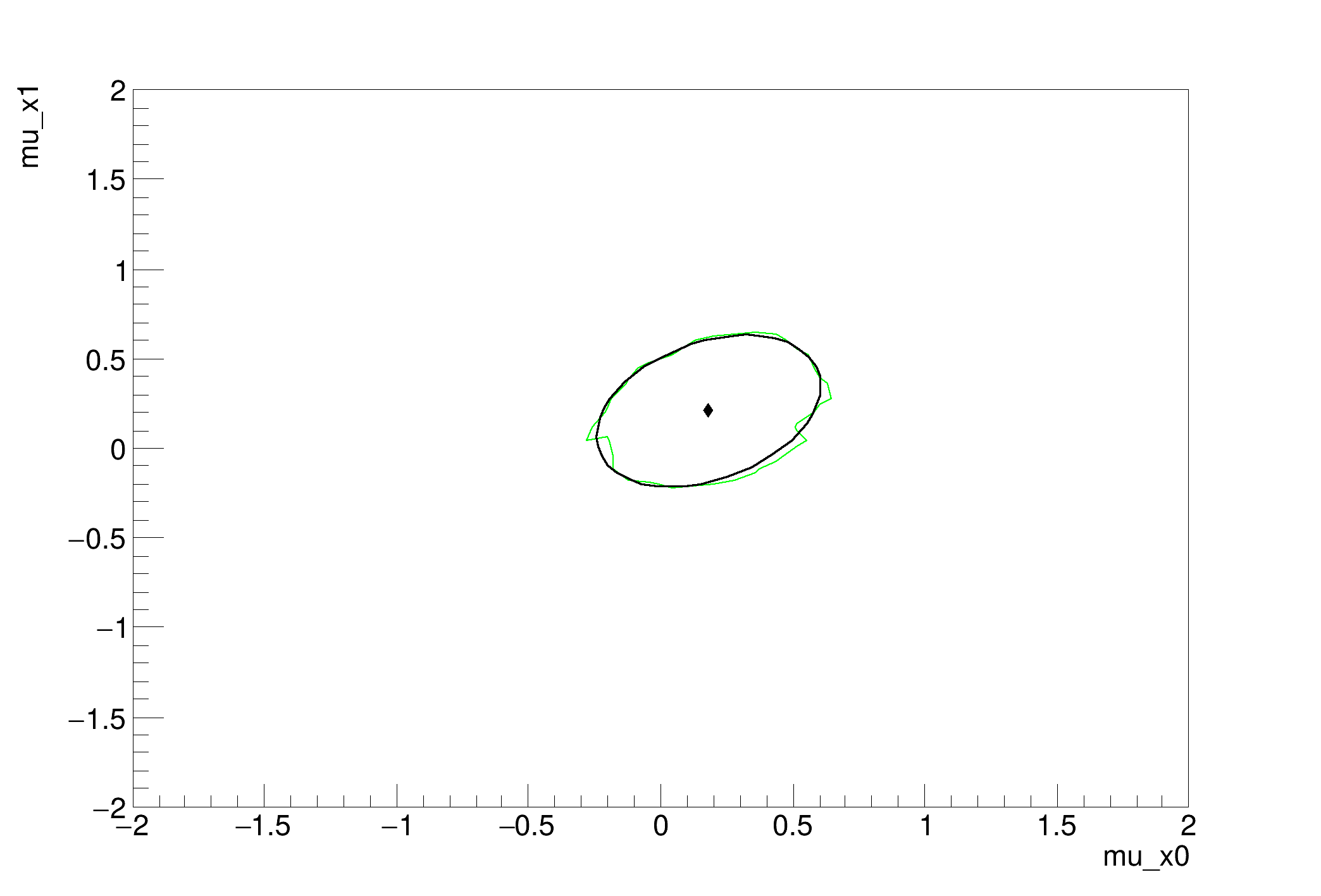
We use this mainly as a test of Markov Chain Monte Carlo and we compare the result to the profile likelihood ratio.
We use the proposal helper to create a customized proposal function for this problem.
For N=4 and 2 parameters of interest it takes about 10-20 seconds and the acceptance rate is 37%
[#1] INFO:Fitting -- RooAbsPdf::fitTo(mvg_over_mvg_Int[x0,x1,x2,x3]) fixing normalization set for coefficient determination to observables in data
[#1] INFO:Fitting -- using CPU computation library compiled with -mavx512
[#1] INFO:Fitting -- RooAddition::defaultErrorLevel(nll_mvg_over_mvg_Int[x0,x1,x2,x3]_mvgData) Summation contains a RooNLLVar, using its error level
[#1] INFO:Minimization -- RooAbsMinimizerFcn::setOptimizeConst: activating const optimization
Minuit2Minimizer: Minimize with max-calls 2000 convergence for edm < 1 strategy 1
Minuit2Minimizer : Valid minimum - status = 0
FVAL = 706.560063684865781
Edm = 9.79361278577427602e-06
Nfcn = 68
mu_x0 = 0.180728 +/- 0.17298 (limited)
mu_x1 = 0.207351 +/- 0.172978 (limited)
mu_x2 = -0.0159412 +/- 0.172984 (limited)
mu_x3 = 0.12343 +/- 0.172982 (limited)
[#1] INFO:Minimization -- RooAbsMinimizerFcn::setOptimizeConst: deactivating const optimization
[#1] INFO:Fitting -- RooAbsPdf::fitTo(mvg_over_mvg_Int[x0,x1,x2,x3]) fixing normalization set for coefficient determination to observables in data
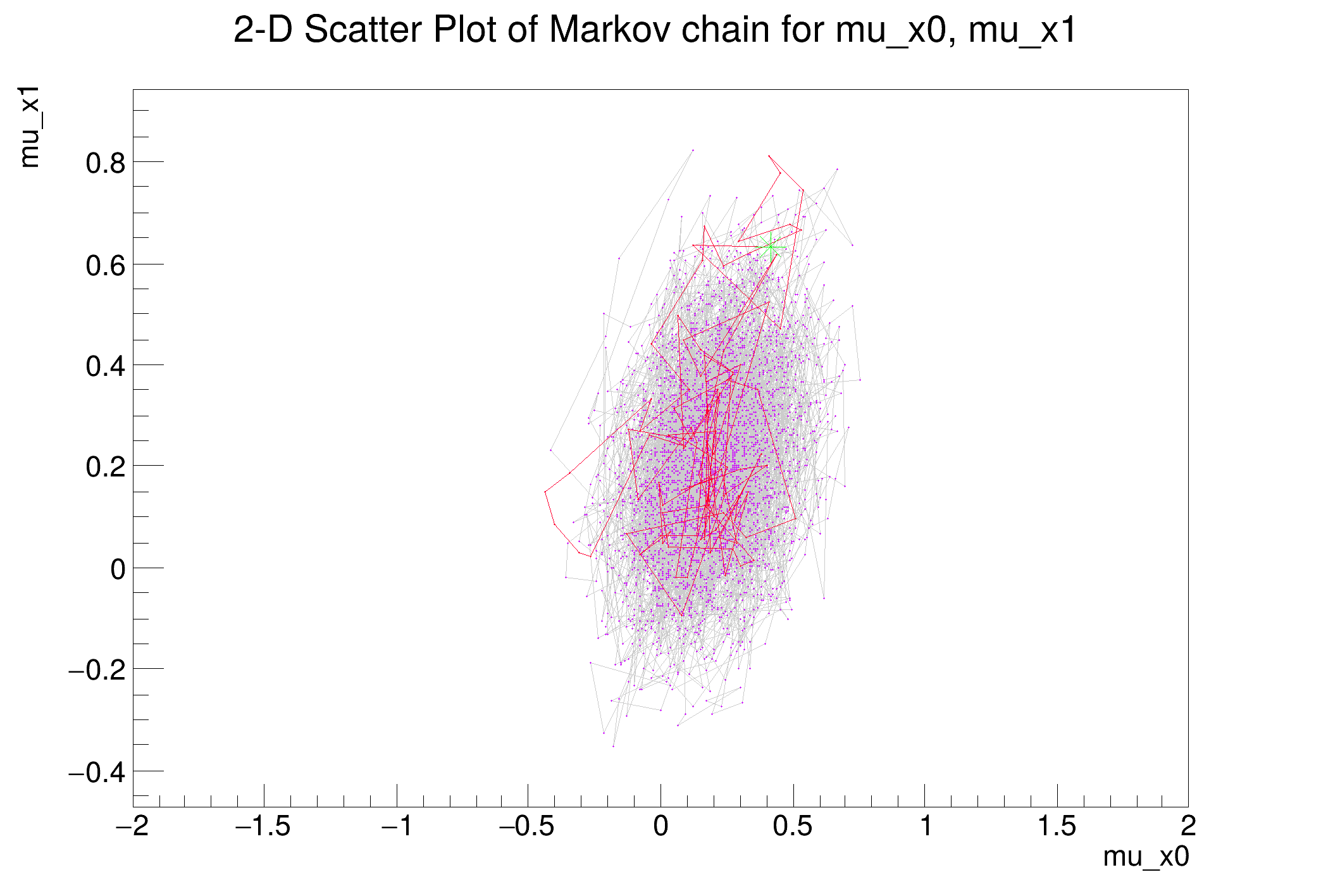
Metropolis-Hastings progress: ....................................................................................................
[#1] INFO:Eval -- Proposal acceptance rate: 37.1%
[#1] INFO:Eval -- Number of steps in chain: 3710
[#1] INFO:InputArguments -- The deprecated RooFit::CloneData(1) option passed to createNLL() is ignored.
[#1] INFO:Fitting -- RooAbsPdf::fitTo(mvg_over_mvg_Int[x0,x1,x2,x3]) fixing normalization set for coefficient determination to observables in data
[#0] PROGRESS:Minimization -- ProfileLikelihoodCalcultor::DoGLobalFit - find MLE
[#1] INFO:Fitting -- RooAddition::defaultErrorLevel(nll_mvg_over_mvg_Int[x0,x1,x2,x3]_mvgData) Summation contains a RooNLLVar, using its error level
[#1] INFO:Minimization -- RooAbsMinimizerFcn::setOptimizeConst: activating const optimization
[#0] PROGRESS:Minimization -- ProfileLikelihoodCalcultor::DoMinimizeNLL - using Minuit2 / Migrad with strategy 1
[#1] INFO:Minimization --
RooFitResult: minimized FCN value: 706.56, estimated distance to minimum: 3.96149e-12
covariance matrix quality: Full, accurate covariance matrix
Status : MINIMIZE=0
Floating Parameter FinalValue +/- Error
-------------------- --------------------------
mu_x0 1.8118e-01 +/- 1.73e-01
mu_x1 2.0792e-01 +/- 1.73e-01
mu_x2 -1.6067e-02 +/- 1.73e-01
mu_x3 1.2371e-01 +/- 1.73e-01
[#1] INFO:Minimization -- RooProfileLL::evaluate(RooEvaluatorWrapper_Profile[mu_x0,mu_x1]) Creating instance of MINUIT
[#1] INFO:Fitting -- RooAddition::defaultErrorLevel(nll_mvg_over_mvg_Int[x0,x1,x2,x3]_mvgData) Summation contains a RooNLLVar, using its error level
[#1] INFO:Minimization -- RooProfileLL::evaluate(RooEvaluatorWrapper_Profile[mu_x0,mu_x1]) determining minimum likelihood for current configurations w.r.t all observable
[#1] INFO:Minimization -- RooProfileLL::evaluate(RooEvaluatorWrapper_Profile[mu_x0,mu_x1]) minimum found at (mu_x0=0.181184, mu_x1=0.207918)
..[#1] INFO:Minimization -- LikelihoodInterval - Finding the contour of mu_x0 ( 0 ) and mu_x1 ( 1 )
MCMC interval on p0: [-0.28, 0.6]
MCMC interval on p1: [-0.2, 0.6]
Real time 0:00:00, CP time 0.610
#include <cstdlib>
using std::cout, std::endl;
{
for (i = 0; i < dim; i++) {
}
for (i = 0; i <
nPOI; i++) {
}
for (i = 0; i < dim; i++) {
for (
j = 0;
j < dim;
j++) {
else
}
}
std::unique_ptr<RooDataSet>
data{
mvg.generate(xVec, 100)};
ph.SetCovMatrix(
fit->covarianceMatrix());
ph.SetUpdateProposalParameters(
true);
mc.SetConfidenceLevel(0.95);
mc.SetNumBurnInSteps(100);
plc.SetConfidenceLevel(0.95);
cout <<
"MCMC interval: [" << ll <<
", " <<
ul <<
"]" << endl;
}
cout <<
"MCMC interval on p0: [" << ll <<
", " <<
ul <<
"]" << endl;
cout <<
"MCMC interval on p1: [" << ll <<
", " <<
ul <<
"]" << endl;
}
}
ROOT::Detail::TRangeCast< T, true > TRangeDynCast
TRangeDynCast is an adapter class that allows the typed iteration through a TCollection.
winID h TVirtualViewer3D TVirtualGLPainter p
Option_t Option_t TPoint TPoint const char GetTextMagnitude GetFillStyle GetLineColor GetLineWidth GetMarkerStyle GetTextAlign GetTextColor GetTextSize void data
char * Form(const char *fmt,...)
Formats a string in a circular formatting buffer.
virtual bool add(const RooAbsArg &var, bool silent=false)
Add the specified argument to list.
RooArgList is a container object that can hold multiple RooAbsArg objects.
RooAbsArg * at(Int_t idx) const
Return object at given index, or nullptr if index is out of range.
RooArgSet is a container object that can hold multiple RooAbsArg objects.
Multivariate Gaussian p.d.f.
Variable that can be changed from the outside.
This class provides simple and straightforward utilities to plot a LikelihoodInterval object.
LikelihoodInterval is a concrete implementation of the RooStats::ConfInterval interface.
Bayesian Calculator estimating an interval or a credible region using the Markov-Chain Monte Carlo me...
This class provides simple and straightforward utilities to plot a MCMCInterval object.
MCMCInterval is a concrete implementation of the RooStats::ConfInterval interface.
ModelConfig is a simple class that holds configuration information specifying how a model should be u...
The ProfileLikelihoodCalculator is a concrete implementation of CombinedCalculator (the interface cla...
ProposalFunction is an interface for all proposal functions that would be used with a Markov Chain Mo...
Persistable container for RooFit projects.
void Start(Bool_t reset=kTRUE)
Start the stopwatch.
void Print(Option_t *option="") const override
Print the real and cpu time passed between the start and stop events.
RooCmdArg Save(bool flag=true)
fit(model, train_loader, val_loader, num_epochs, batch_size, optimizer, criterion, save_best, scheduler)
The namespace RooFit contains mostly switches that change the behaviour of functions of PDFs (or othe...
Namespace for the RooStats classes.