

This example is a generalization of the on/off problem.
This example is a generalization of the on/off problem. It's a common setup for SUSY searches. Imagine that one has two variables "x" and "y" (eg. missing ET and SumET), see figure. The signal region has high values of both of these variables (top right). One can see low values of "x" or "y" acting as side-bands. If we just used "y" as a sideband, we would have the on/off problem.
- In the signal region we observe non events and expect s+b events.
- In the region with low values of "y" (bottom right) we observe noff events and expect tau*b events. Note the significance of tau. In the background only case:
Option_t Option_t TPoint TPoint const char GetTextMagnitude GetFillStyle GetLineColor GetLineWidth GetMarkerStyle GetTextAlign GetTextColor GetTextSize void on
If tau is known, this model is sufficient, but often tau is not known exactly. So one can use low values of "x" as an additional constraint for tau. Note that this technique critically depends on the notion that the joint distribution for "x" and "y" can be factorized. Generally, these regions have many events, so it the ratio can be measured very precisely there. So we extend the model to describe the left two boxes... denoted with "bar".
- In the upper left we observe nonbar events and expect bbar events
- In the bottom left we observe noffbar events and expect tau bbar events Note again we have:
One can further expand the model to account for the systematic associated to assuming the distribution of "x" and "y" factorizes (eg. that tau is the same for off/on and offbar/onbar). This can be done in several ways, but here we introduce an additional parameter rho, which so that one set of models will use tau and the other tau*rho. The choice is arbitrary, but it has consequences on the numerical stability of the algorithms. The "bar" measurements typically have more events (& smaller relative errors). If we choose
the product tau*rho will be known very precisely (~1/sqrt(bbar)) and the contour in those parameters will be narrow and have a non-trivial tau~1/rho shape. However, if we choose to put rho on the non/noff measurements (where the product will have an error ~1/sqrt(b)), the contours will be more amenable to numerical techniques. Thus, here we choose to define:
|
|---------------------------+
| | |
| | |
|---------------+-----------|
| | |
| | |
+----------------------------->
x Left in this way, the problem is under-constrained. However, one may have some auxiliary measurement (usually based on Monte Carlo) to constrain rho. Let us call this auxiliary measurement that gives the nominal value of rho "rhonom". Thus, there is a 'constraint' term in the full model: P(rhonom | rho). In this case, we consider a Gaussian constraint with standard deviation sigma.
In the example, the initial values of the parameters are:
- s = 40
- tau = 5
- rho = 1
and in the toy dataset:
Note, the covariance matrix of the parameters has large off-diagonal terms. Clearly s,b are anti-correlated. Similarly, since noffbar >> nonbar, one would expect bbar,tau to be anti-correlated.
This can be seen below.
b 0.96820 1.000 0.191 -0.942 -0.762 -0.209
bbar 0.91191 0.191 1.000 0.000 -0.146 -0.912
rho 0.96348 -0.942 0.000 1.000 0.718 -0.000
s 0.76250 -0.762 -0.146 0.718 1.000 0.160
tau 0.92084 -0.209 -0.912 -0.000 0.160 1.000
Similarly, since tau*rho appears as a product, we expect rho,tau to be anti-correlated. When the error on rho is significantly larger than 1/sqrt(bbar), tau is essentially known and the correlation is minimal (tau mainly cares about bbar, and rho about b,s). In the alternate parametrization (bbar* tau * rho) the correlation coefficient for rho,tau is large (and negative).
The code below uses best-practices for RooFit & RooStats as of June 2010.
It proceeds as follows:
- create a workspace to hold the model
- use workspace factory to quickly create the terms of the model
- use workspace factory to define total model (a prod pdf)
- create a RooStats ModelConfig to specify observables, parameters of interest
- add to the ModelConfig a prior on the parameters for Bayesian techniques note, the pdf it is factorized for parameters of interest & nuisance params
- visualize the model
- write the workspace to a file
- use several of RooStats IntervalCalculators & compare results
[#0] WARNING:InputArguments -- The parameter 'sigma' with range [-inf, inf] of the RooGaussian 'mcCons' exceeds the safe range of (0, inf). Advise to limit its range.
[#1] INFO:ObjectHandling -- RooWorkspace::import(wspace) importing dataset modelData
[#1] INFO:InputArguments -- The deprecated RooFit::CloneData(1) option passed to createNLL() is ignored.
[#1] INFO:Fitting -- RooAbsPdf::fitTo(model) fixing normalization set for coefficient determination to observables in data
[#1] INFO:Fitting -- using CPU computation library compiled with -mavx512
[#0] PROGRESS:Minimization -- ProfileLikelihoodCalcultor::DoGLobalFit - find MLE
[#1] INFO:Fitting -- RooAddition::defaultErrorLevel(nll_model_modelData) Summation contains a RooNLLVar, using its error level
[#1] INFO:Minimization -- RooAbsMinimizerFcn::setOptimizeConst: activating const optimization
[#0] PROGRESS:Minimization -- ProfileLikelihoodCalcultor::DoMinimizeNLL - using Minuit2 / with strategy 1
[#1] INFO:Minimization --
RooFitResult: minimized FCN value: 16.2872, estimated distance to minimum: 3.94032e-11
covariance matrix quality: Full, accurate covariance matrix
Status : MINIMIZE=0
Floating Parameter FinalValue +/- Error
-------------------- --------------------------
b 8.3599e+01 +/- 1.39e+01
bbar 9.9300e+02 +/- 3.15e+01
rho 1.2784e+00 +/- 1.99e-01
s 5.5401e+01 +/- 1.78e+01
tau 4.9406e+00 +/- 1.72e-01
Bayesian Calc. only supports on parameter of interest
[#1] INFO:Fitting -- RooAbsPdf::fitTo(model) fixing normalization set for coefficient determination to observables in data
[#1] INFO:Fitting -- RooAddition::defaultErrorLevel(nll_model_modelData) Summation contains a RooNLLVar, using its error level
[#1] INFO:Minimization -- RooAbsMinimizerFcn::setOptimizeConst: activating const optimization
Minuit2Minimizer: Minimize with max-calls 2500 convergence for edm < 1 strategy 1
Minuit2Minimizer : Valid minimum - status = 0
FVAL = 16.2877174059493548
Edm = 0.000500993073975738328
Nfcn = 161
b = 83.6236 +/- 13.8841 (limited)
bbar = 992.063 +/- 31.4891 (limited)
rho = 1.27663 +/- 0.198616 (limited)
s = 55.4223 +/- 17.8332 (limited)
tau = 4.94561 +/- 0.171974 (limited)
[#1] INFO:Minimization -- RooAbsMinimizerFcn::setOptimizeConst: deactivating const optimization
[#1] INFO:Minimization -- RooProfileLL::evaluate(RooEvaluatorWrapper_Profile[s]) Creating instance of MINUIT
[#1] INFO:Fitting -- RooAddition::defaultErrorLevel(nll_model_modelData) Summation contains a RooNLLVar, using its error level
[#1] INFO:Minimization -- RooProfileLL::evaluate(RooEvaluatorWrapper_Profile[s]) determining minimum likelihood for current configurations w.r.t all observable
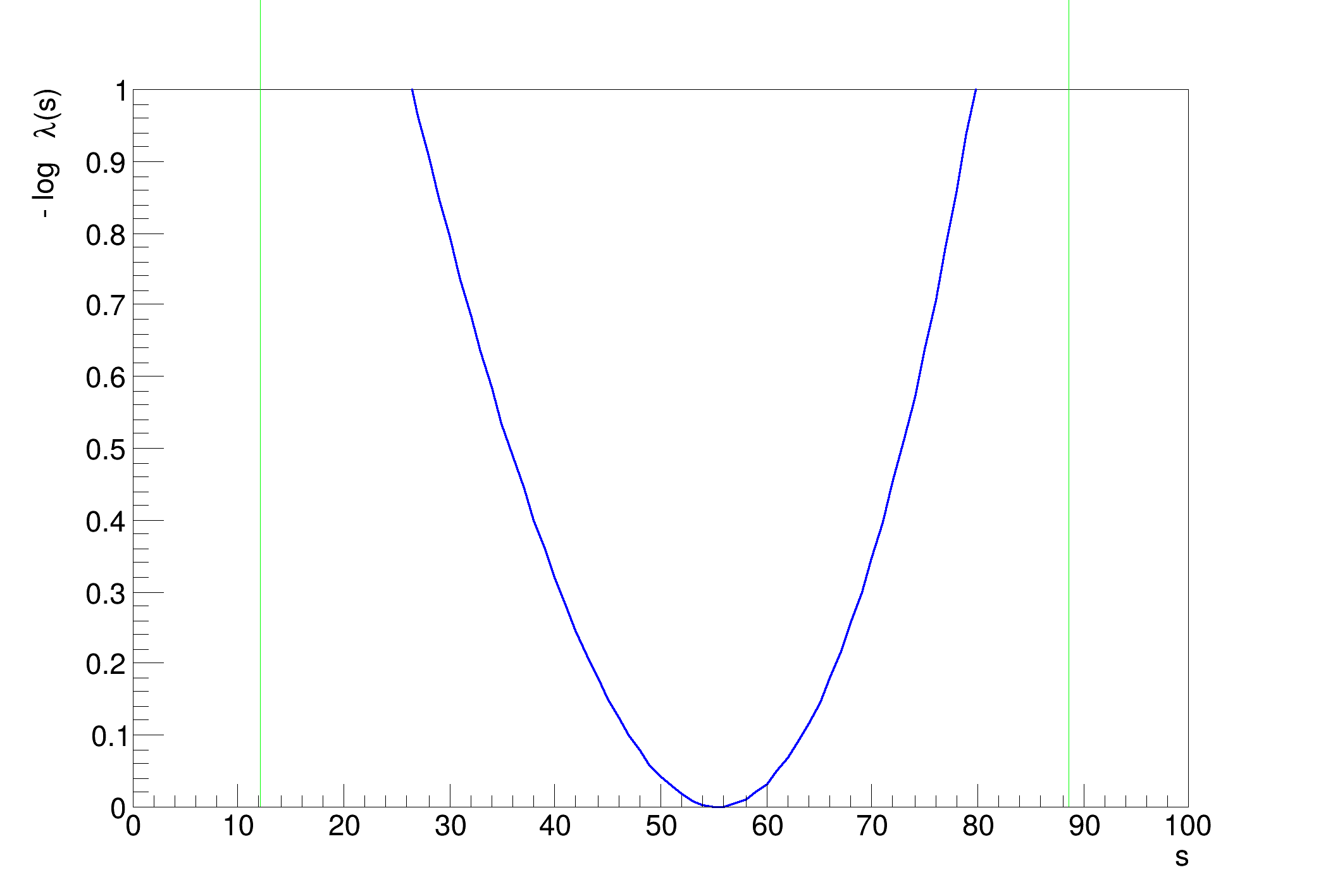
[#1] INFO:Minimization -- RooProfileLL::evaluate(RooEvaluatorWrapper_Profile[s]) minimum found at (s=55.3131)
.
[#1] INFO:Minimization -- RooProfileLL::evaluate(RooEvaluatorWrapper_Profile[s]) Creating instance of MINUIT
[#1] INFO:Fitting -- RooAddition::defaultErrorLevel(nll_model_modelData) Summation contains a RooNLLVar, using its error level
[#1] INFO:Minimization -- RooProfileLL::evaluate(RooEvaluatorWrapper_Profile[s]) determining minimum likelihood for current configurations w.r.t all observable
[#0] ERROR:InputArguments -- RooArgSet::checkForDup: ERROR argument with name s is already in this set
[#1] INFO:Minimization -- RooProfileLL::evaluate(RooEvaluatorWrapper_Profile[s]) minimum found at (s=55.388)
..........................................................................................................................................................................................................Profile Likelihood interval on s = [12.1902, 88.6871]
Real time 0:00:00, CP time 0.130
{
wspace->
factory(
"Poisson::on(non[0,1000], sum::splusb(s[40,0,100],b[100,0,300]))");
wspace->
factory(
"Poisson::off(noff[0,5000], prod::taub(b,tau[5,3,7],rho[1,0,2]))");
wspace->
factory(
"Poisson::onbar(nonbar[0,10000], bbar[1000,500,2000])");
wspace->
factory(
"Poisson::offbar(noffbar[0,1000000], prod::lambdaoffbar(bbar, tau))");
wspace->
factory(
"Gaussian::mcCons(rhonom[1.,0,2], rho, sigma[.2])");
wspace->
factory(
"PROD::model(on,off,onbar,offbar,mcCons)");
wspace->
defineSet(
"obs",
"non,noff,nonbar,noffbar,rhonom");
wspace->
factory(
"Uniform::prior_poi({s})");
wspace->
factory(
"Uniform::prior_nuis({b,bbar,tau, rho})");
wspace->
factory(
"PROD::prior(prior_poi,prior_nuis)");
plc.SetConfidenceLevel(0.95);
fc.SetConfidenceLevel(0.95);
fc.FluctuateNumDataEntries(
false);
fc.UseAdaptiveSampling(
true);
}
bc.SetConfidenceLevel(0.95);
} else {
cout << "Bayesian Calc. only supports on parameter of interest" << endl;
}
ph.SetCovMatrix(
fit->covarianceMatrix());
ph.SetUpdateProposalParameters(
kTRUE);
mc.SetConfidenceLevel(0.95);
mc.SetProposalFunction(*
pf);
mc.SetNumBurnInSteps(500);
mc.SetLeftSideTailFraction(0.5);
}
bc.SetScanOfPosterior(20);
}
else
}
cout <<
"Profile Likelihood interval on s = [" <<
plInt->LowerLimit(*wspace->
var(
"s")) <<
", " <<
plInt->UpperLimit(*wspace->
var(
"s")) <<
"]" << endl;
cout <<
"Bayesian interval on s = [" <<
bInt->LowerLimit() <<
", " <<
bInt->UpperLimit() <<
"]" << endl;
}
cout <<
"Feldman Cousins interval on s = [" <<
fcInt->LowerLimit(*wspace->
var(
"s")) <<
", " <<
fcInt->UpperLimit(*wspace->
var(
"s")) <<
"]" << endl;
}
cout <<
"MCMC interval on s = [" <<
mcInt->LowerLimit(*wspace->
var(
"s")) <<
", " <<
mcInt->UpperLimit(*wspace->
var(
"s")) <<
"]" << endl;
}
}
ROOT::Detail::TRangeCast< T, true > TRangeDynCast
TRangeDynCast is an adapter class that allows the typed iteration through a TCollection.
Option_t Option_t TPoint TPoint const char GetTextMagnitude GetFillStyle GetLineColor GetLineWidth GetMarkerStyle GetTextAlign GetTextColor GetTextSize void data
RooFit::OwningPtr< RooFitResult > fitTo(RooAbsData &data, CmdArgs_t const &... cmdArgs)
Fit PDF to given dataset.
RooFit::OwningPtr< RooDataSet > generate(const RooArgSet &whatVars, Int_t nEvents, const RooCmdArg &arg1, const RooCmdArg &arg2={}, const RooCmdArg &arg3={}, const RooCmdArg &arg4={}, const RooCmdArg &arg5={})
See RooAbsPdf::generate(const RooArgSet&,const RooCmdArg&,const RooCmdArg&,const RooCmdArg&,...
RooArgSet is a container object that can hold multiple RooAbsArg objects.
static RooMsgService & instance()
Return reference to singleton instance.
Plot frame and a container for graphics objects within that frame.
static TRandom * randomGenerator()
Return a pointer to a singleton random-number generator implementation.
BayesianCalculator is a concrete implementation of IntervalCalculator, providing the computation of a...
The FeldmanCousins class (like the Feldman-Cousins technique) is essentially a specific configuration...
This class provides simple and straightforward utilities to plot a LikelihoodInterval object.
LikelihoodInterval is a concrete implementation of the RooStats::ConfInterval interface.
Bayesian Calculator estimating an interval or a credible region using the Markov-Chain Monte Carlo me...
This class provides simple and straightforward utilities to plot a MCMCInterval object.
MCMCInterval is a concrete implementation of the RooStats::ConfInterval interface.
ModelConfig is a simple class that holds configuration information specifying how a model should be u...
PointSetInterval is a concrete implementation of the ConfInterval interface.
The ProfileLikelihoodCalculator is a concrete implementation of CombinedCalculator (the interface cla...
ProposalFunction is an interface for all proposal functions that would be used with a Markov Chain Mo...
SimpleInterval is a concrete implementation of the ConfInterval interface.
Persistable container for RooFit projects.
RooAbsPdf * pdf(RooStringView name) const
Retrieve p.d.f (RooAbsPdf) with given name. A null pointer is returned if not found.
const RooArgSet * set(RooStringView name)
Return pointer to previously defined named set with given nmame If no such set is found a null pointe...
bool import(const RooAbsArg &arg, const RooCmdArg &arg1={}, const RooCmdArg &arg2={}, const RooCmdArg &arg3={}, const RooCmdArg &arg4={}, const RooCmdArg &arg5={}, const RooCmdArg &arg6={}, const RooCmdArg &arg7={}, const RooCmdArg &arg8={}, const RooCmdArg &arg9={})
Import a RooAbsArg object, e.g.
RooFactoryWSTool & factory()
Return instance to factory tool.
RooRealVar * var(RooStringView name) const
Retrieve real-valued variable (RooRealVar) with given name. A null pointer is returned if not found.
bool defineSet(const char *name, const RooArgSet &aset, bool importMissing=false)
Define a named RooArgSet with given constituents.
void Start(Bool_t reset=kTRUE)
Start the stopwatch.
void Print(Option_t *option="") const override
Print the real and cpu time passed between the start and stop events.
RooCmdArg Save(bool flag=true)
fit(model, train_loader, val_loader, num_epochs, batch_size, optimizer, criterion, save_best, scheduler)
The namespace RooFit contains mostly switches that change the behaviour of functions of PDFs (or othe...
MsgLevel
Verbosity level for RooMsgService::StreamConfig in RooMsgService.
Namespace for the RooStats classes.
- Authors
- authors: Kyle Cranmer, Tanja Rommerskirchen
Definition in file FourBinInstructional.C.


