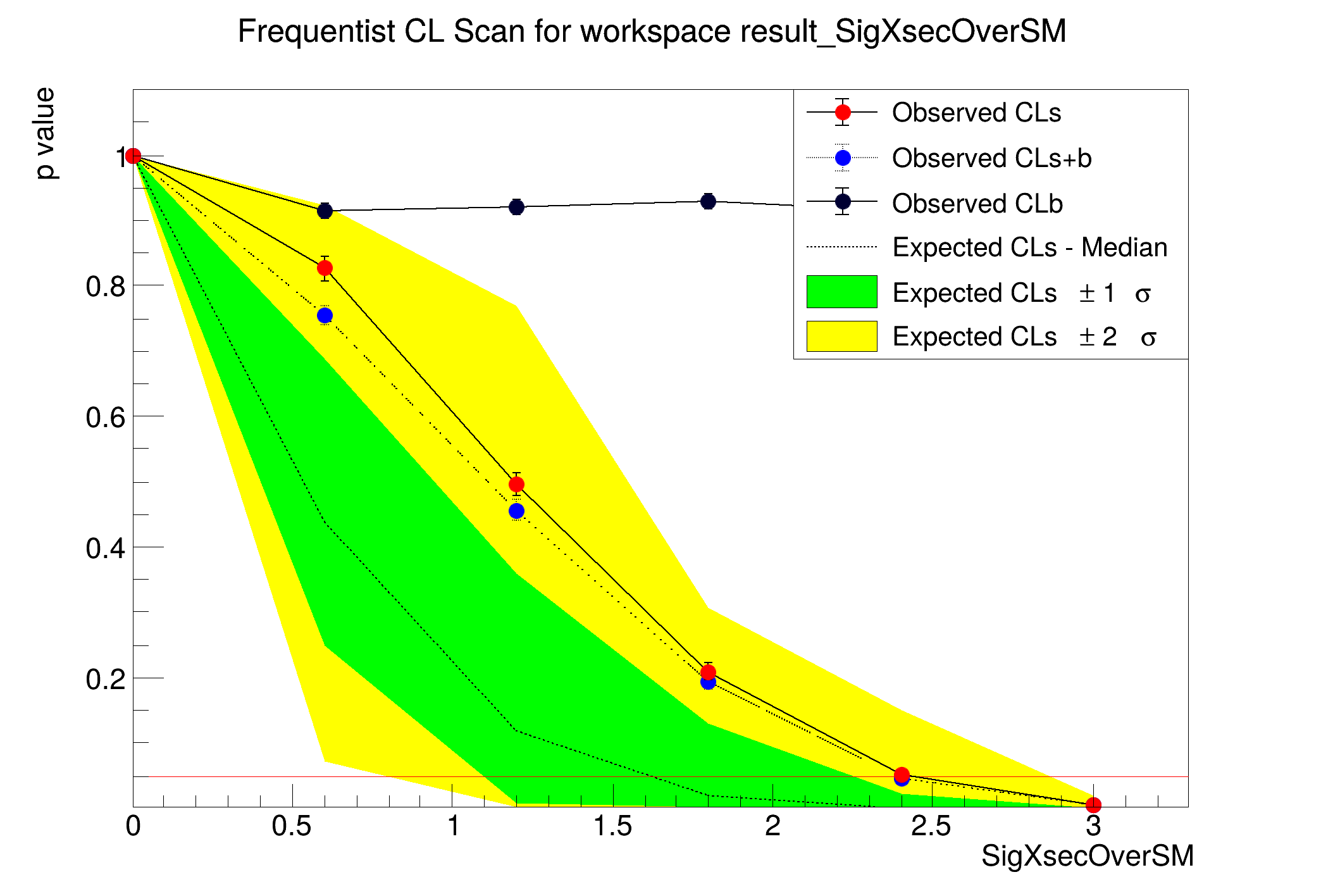
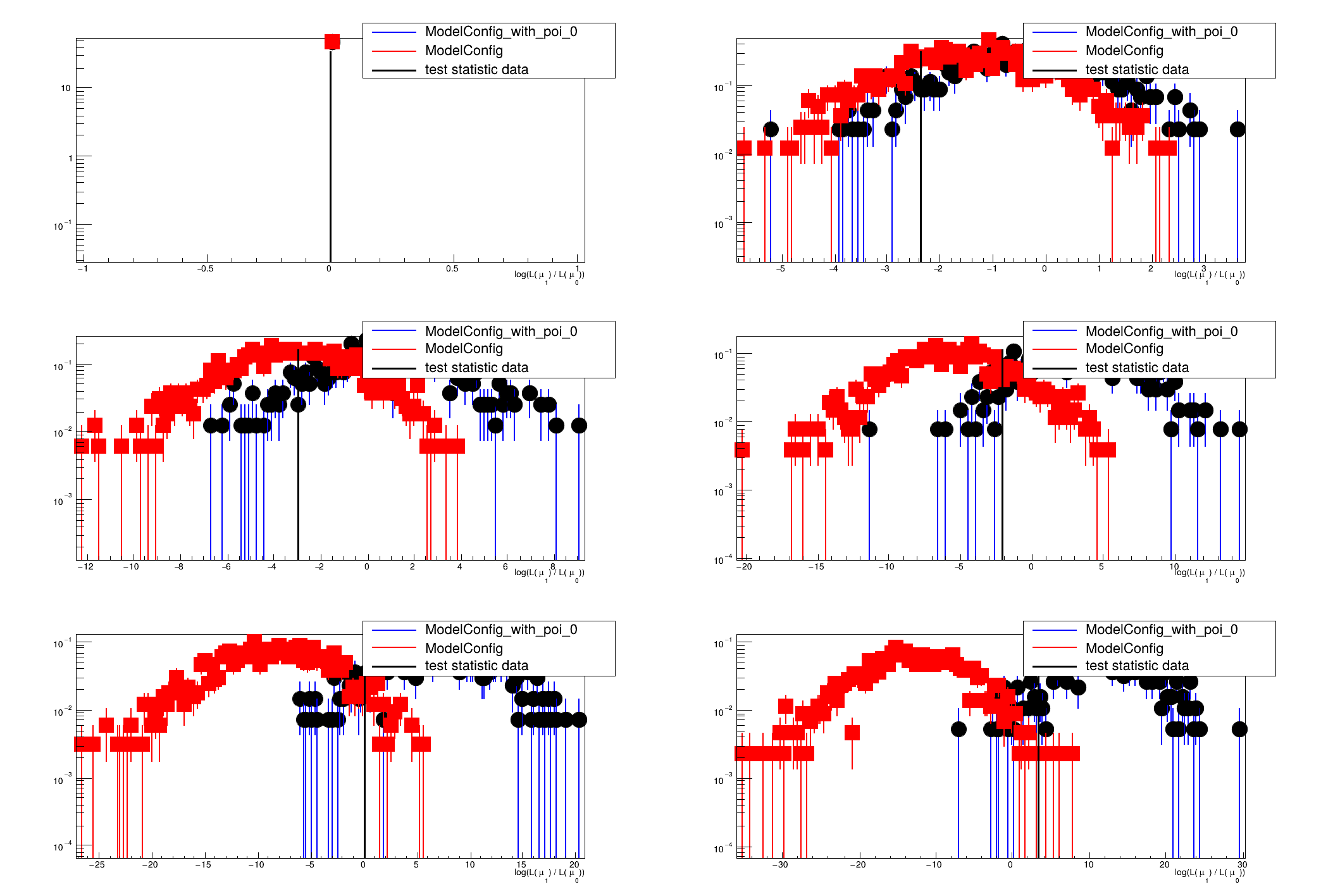
This macro will perform a scan of the p-values for computing the interval or limit
0x8945870 results/example_combined_GaussExample_model.root
Running HypoTestInverter on the workspace combined
RooWorkspace(combined) combined contents
variables
---------
(Lumi,SigXsecOverSM,alpha_syst1,alpha_syst2,alpha_syst3,channelCat,gamma_stat_channel1_bin_0,gamma_stat_channel1_bin_1,nom_alpha_syst1,nom_alpha_syst2,nom_alpha_syst3,nom_gamma_stat_channel1_bin_0,nom_gamma_stat_channel1_bin_1,nominalLumi,obs_x_channel1)
p.d.f.s
-------
RooGaussian::alpha_syst1Constraint[ x=alpha_syst1 mean=nom_alpha_syst1 sigma=1 ] = 1
RooGaussian::alpha_syst2Constraint[ x=alpha_syst2 mean=nom_alpha_syst2 sigma=1 ] = 1
RooGaussian::alpha_syst3Constraint[ x=alpha_syst3 mean=nom_alpha_syst3 sigma=1 ] = 1
RooRealSumPdf::channel1_model[ signal_channel1_scaleFactors * signal_channel1_shapes + background1_channel1_scaleFactors * background1_channel1_shapes + background2_channel1_scaleFactors * background2_channel1_shapes ] = 240
RooPoisson::gamma_stat_channel1_bin_0_constraint[ x=nom_gamma_stat_channel1_bin_0 mean=gamma_stat_channel1_bin_0_poisMean ] = 0.019943
RooPoisson::gamma_stat_channel1_bin_1_constraint[ x=nom_gamma_stat_channel1_bin_1 mean=gamma_stat_channel1_bin_1_poisMean ] = 0.039861
RooGaussian::lumiConstraint[ x=Lumi mean=nominalLumi sigma=0.1 ] = 1
RooProdPdf::model_channel1[ lumiConstraint * alpha_syst1Constraint * alpha_syst2Constraint * alpha_syst3Constraint * gamma_stat_channel1_bin_0_constraint * gamma_stat_channel1_bin_1_constraint * channel1_model(obs_x_channel1) ] = 0.190787
RooSimultaneous::simPdf[ indexCat=channelCat channel1=model_channel1 ] = 0.190787
functions
--------
RooHistFunc::background1_channel1_Hist_alphanominal[ depList=(obs_x_channel1) depList=(obs_x_channel1) ] = 100
RooStats::HistFactory::FlexibleInterpVar::background1_channel1_epsilon[ paramList=(alpha_syst2) ] = 1
RooProduct::background1_channel1_scaleFactors[ background1_channel1_epsilon * Lumi ] = 1
RooProduct::background1_channel1_shapes[ background1_channel1_Hist_alphanominal * mc_stat_channel1 * channel1_model_binWidth ] = 200
RooHistFunc::background2_channel1_Hist_alphanominal[ depList=(obs_x_channel1) depList=(obs_x_channel1) ] = 0
RooStats::HistFactory::FlexibleInterpVar::background2_channel1_epsilon[ paramList=(alpha_syst3) ] = 1
RooProduct::background2_channel1_scaleFactors[ background2_channel1_epsilon * Lumi ] = 1
RooProduct::background2_channel1_shapes[ background2_channel1_Hist_alphanominal * mc_stat_channel1 * channel1_model_binWidth ] = 0
RooBinWidthFunction::channel1_model_binWidth[ HistFuncForBinWidth=signal_channel1_Hist_alphanominal HistFuncForBinWidth=signal_channel1_Hist_alphanominal ] = 2
RooProduct::gamma_stat_channel1_bin_0_poisMean[ gamma_stat_channel1_bin_0 * gamma_stat_channel1_bin_0_tau ] = 400
RooProduct::gamma_stat_channel1_bin_1_poisMean[ gamma_stat_channel1_bin_1 * gamma_stat_channel1_bin_1_tau ] = 100
ParamHistFunc::mc_stat_channel1[ ] = 1
RooHistFunc::signal_channel1_Hist_alphanominal[ depList=(obs_x_channel1) depList=(obs_x_channel1) ] = 20
RooStats::HistFactory::FlexibleInterpVar::signal_channel1_epsilon[ paramList=(alpha_syst1) ] = 1
RooProduct::signal_channel1_scaleFactors[ signal_channel1_epsilon * SigXsecOverSM * Lumi ] = 1
RooProduct::signal_channel1_shapes[ signal_channel1_Hist_alphanominal * channel1_model_binWidth ] = 40
datasets
--------
RooDataSet::obsData(obs_x_channel1,channelCat)
RooDataSet::asimovData(obs_x_channel1,channelCat)
embedded datasets (in pdfs and functions)
-----------------------------------------
RooDataHist::signal_channel1_Hist_alphanominalDHist(obs_x_channel1)
RooDataHist::background1_channel1_Hist_alphanominalDHist(obs_x_channel1)
RooDataHist::background2_channel1_Hist_alphanominalDHist(obs_x_channel1)
parameter snapshots
-------------------
NominalParamValues = (nominalLumi=1[C],nom_alpha_syst1=0[C],nom_alpha_syst2=0[C],nom_alpha_syst3=0[C],nom_gamma_stat_channel1_bin_0=400[C],nom_gamma_stat_channel1_bin_1=100[C],obs_x_channel1=1.25,Lumi=1 +/- 0.1[C],alpha_syst1=0 +/- 1[C],alpha_syst2=0 +/- 1,alpha_syst3=0 +/- 1,gamma_stat_channel1_bin_0=1 +/- 0.05,gamma_stat_channel1_bin_1=1 +/- 0.1,SigXsecOverSM=1)
named sets
----------
ModelConfig_GlobalObservables:(nominalLumi,nom_alpha_syst1,nom_alpha_syst2,nom_alpha_syst3,nom_gamma_stat_channel1_bin_0,nom_gamma_stat_channel1_bin_1)
ModelConfig_NuisParams:(alpha_syst2,alpha_syst3,gamma_stat_channel1_bin_0,gamma_stat_channel1_bin_1)
ModelConfig_Observables:(obs_x_channel1,channelCat)
ModelConfig_POI:(SigXsecOverSM)
globalObservables:(nominalLumi,nom_alpha_syst1,nom_alpha_syst2,nom_alpha_syst3,nom_gamma_stat_channel1_bin_0,nom_gamma_stat_channel1_bin_1)
observables:(obs_x_channel1,channelCat)
generic objects
---------------
RooStats::ModelConfig::ModelConfig
Using data set obsData
StandardHypoTestInvDemo : POI initial value: SigXsecOverSM = 1
[#1] INFO:InputArguments -- HypoTestInverter ---- Input models:
using as S+B (null) model : ModelConfig
using as B (alternate) model : ModelConfig_with_poi_0
Doing a fixed scan in interval : 0 , 5
[#1] INFO:Eval -- HypoTestInverter::GetInterval - run a fixed scan
[#0] WARNING:InputArguments -- HypoTestInverter::RunFixedScan - xMax > upper bound, using xmax = 3
[#1] INFO:ObjectHandling -- RooWorkspace::saveSnapshot(combined) replacing previous snapshot with name ModelConfig__snapshot
[#0] PROGRESS:Eval -- Running for SigXsecOverSM = 0
=== Using the following for ModelConfig ===
Observables: RooArgSet:: = (obs_x_channel1,channelCat)
Parameters of Interest: RooArgSet:: = (SigXsecOverSM)
Nuisance Parameters: RooArgSet:: = (alpha_syst2,alpha_syst3,gamma_stat_channel1_bin_0,gamma_stat_channel1_bin_1)
Global Observables: RooArgSet:: = (nominalLumi,nom_alpha_syst1,nom_alpha_syst2,nom_alpha_syst3,nom_gamma_stat_channel1_bin_0,nom_gamma_stat_channel1_bin_1)
PDF: RooSimultaneous::simPdf[ indexCat=channelCat channel1=model_channel1 ] = 0.158989
Snapshot:
1) 0x9866d60 RooRealVar:: SigXsecOverSM = 0 L(0 - 3) "SigXsecOverSM"
=== Using the following for ModelConfig_with_poi_0 ===
Observables: RooArgSet:: = (obs_x_channel1,channelCat)
Parameters of Interest: RooArgSet:: = (SigXsecOverSM)
Nuisance Parameters: RooArgSet:: = (alpha_syst2,alpha_syst3,gamma_stat_channel1_bin_0,gamma_stat_channel1_bin_1)
Global Observables: RooArgSet:: = (nominalLumi,nom_alpha_syst1,nom_alpha_syst2,nom_alpha_syst3,nom_gamma_stat_channel1_bin_0,nom_gamma_stat_channel1_bin_1)
PDF: RooSimultaneous::simPdf[ indexCat=channelCat channel1=model_channel1 ] = 0.158989
Snapshot:
1) 0x9824bd0 RooRealVar:: SigXsecOverSM = 0 L(0 - 3) "SigXsecOverSM"
[#0] PROGRESS:Generation -- Test Statistic on data: 0
[#1] INFO:InputArguments -- Profiling conditional MLEs for Null.
[#1] INFO:InputArguments -- Using a ToyMCSampler. Now configuring for Null.
[#0] PROGRESS:Generation -- generated toys: 500 / 1000
[#1] INFO:InputArguments -- Profiling conditional MLEs for Alt.
[#1] INFO:InputArguments -- Using a ToyMCSampler. Now configuring for Alt.
[#0] PROGRESS:Eval -- P values for SigXsecOverSM = 0
CLs = 1 +/- 0
CLb = 1 +/- 0
CLsplusb = 1 +/- 0
[#1] INFO:ObjectHandling -- RooWorkspace::saveSnapshot(combined) replacing previous snapshot with name ModelConfig__snapshot
[#0] PROGRESS:Eval -- Running for SigXsecOverSM = 0.6
=== Using the following for ModelConfig ===
Observables: RooArgSet:: = (obs_x_channel1,channelCat)
Parameters of Interest: RooArgSet:: = (SigXsecOverSM)
Nuisance Parameters: RooArgSet:: = (alpha_syst2,alpha_syst3,gamma_stat_channel1_bin_0,gamma_stat_channel1_bin_1)
Global Observables: RooArgSet:: = (nominalLumi,nom_alpha_syst1,nom_alpha_syst2,nom_alpha_syst3,nom_gamma_stat_channel1_bin_0,nom_gamma_stat_channel1_bin_1)
PDF: RooSimultaneous::simPdf[ indexCat=channelCat channel1=model_channel1 ] = 0.178068
Snapshot:
1) 0x999bd70 RooRealVar:: SigXsecOverSM = 0.6 L(0 - 3) "SigXsecOverSM"
=== Using the following for ModelConfig_with_poi_0 ===
Observables: RooArgSet:: = (obs_x_channel1,channelCat)
Parameters of Interest: RooArgSet:: = (SigXsecOverSM)
Nuisance Parameters: RooArgSet:: = (alpha_syst2,alpha_syst3,gamma_stat_channel1_bin_0,gamma_stat_channel1_bin_1)
Global Observables: RooArgSet:: = (nominalLumi,nom_alpha_syst1,nom_alpha_syst2,nom_alpha_syst3,nom_gamma_stat_channel1_bin_0,nom_gamma_stat_channel1_bin_1)
PDF: RooSimultaneous::simPdf[ indexCat=channelCat channel1=model_channel1 ] = 0.178068
Snapshot:
1) 0x9864940 RooRealVar:: SigXsecOverSM = 0 L(0 - 3) "SigXsecOverSM"
[#0] PROGRESS:Generation -- Test Statistic on data: -2.35222
[#1] INFO:InputArguments -- Profiling conditional MLEs for Null.
[#1] INFO:InputArguments -- Using a ToyMCSampler. Now configuring for Null.
[#0] PROGRESS:Generation -- generated toys: 500 / 1000
[#1] INFO:InputArguments -- Profiling conditional MLEs for Alt.
[#1] INFO:InputArguments -- Using a ToyMCSampler. Now configuring for Alt.
[#0] PROGRESS:Eval -- P values for SigXsecOverSM = 0.6
CLs = 0.826039 +/- 0.0187037
CLb = 0.914 +/- 0.0125383
CLsplusb = 0.755 +/- 0.0136006
[#1] INFO:ObjectHandling -- RooWorkspace::saveSnapshot(combined) replacing previous snapshot with name ModelConfig__snapshot
[#0] PROGRESS:Eval -- Running for SigXsecOverSM = 1.2
=== Using the following for ModelConfig ===
Observables: RooArgSet:: = (obs_x_channel1,channelCat)
Parameters of Interest: RooArgSet:: = (SigXsecOverSM)
Nuisance Parameters: RooArgSet:: = (alpha_syst2,alpha_syst3,gamma_stat_channel1_bin_0,gamma_stat_channel1_bin_1)
Global Observables: RooArgSet:: = (nominalLumi,nom_alpha_syst1,nom_alpha_syst2,nom_alpha_syst3,nom_gamma_stat_channel1_bin_0,nom_gamma_stat_channel1_bin_1)
PDF: RooSimultaneous::simPdf[ indexCat=channelCat channel1=model_channel1 ] = 0.197147
Snapshot:
1) 0x998c910 RooRealVar:: SigXsecOverSM = 1.2 L(0 - 3) "SigXsecOverSM"
=== Using the following for ModelConfig_with_poi_0 ===
Observables: RooArgSet:: = (obs_x_channel1,channelCat)
Parameters of Interest: RooArgSet:: = (SigXsecOverSM)
Nuisance Parameters: RooArgSet:: = (alpha_syst2,alpha_syst3,gamma_stat_channel1_bin_0,gamma_stat_channel1_bin_1)
Global Observables: RooArgSet:: = (nominalLumi,nom_alpha_syst1,nom_alpha_syst2,nom_alpha_syst3,nom_gamma_stat_channel1_bin_0,nom_gamma_stat_channel1_bin_1)
PDF: RooSimultaneous::simPdf[ indexCat=channelCat channel1=model_channel1 ] = 0.197147
Snapshot:
1) 0x997de10 RooRealVar:: SigXsecOverSM = 0 L(0 - 3) "SigXsecOverSM"
[#0] PROGRESS:Generation -- Test Statistic on data: -2.9364
[#1] INFO:InputArguments -- Profiling conditional MLEs for Null.
[#1] INFO:InputArguments -- Using a ToyMCSampler. Now configuring for Null.
[#0] PROGRESS:Generation -- generated toys: 500 / 1000
[#1] INFO:InputArguments -- Profiling conditional MLEs for Alt.
[#1] INFO:InputArguments -- Using a ToyMCSampler. Now configuring for Alt.
[#0] PROGRESS:Eval -- P values for SigXsecOverSM = 1.2
CLs = 0.495652 +/- 0.018325
CLb = 0.92 +/- 0.0121326
CLsplusb = 0.456 +/- 0.01575
[#1] INFO:ObjectHandling -- RooWorkspace::saveSnapshot(combined) replacing previous snapshot with name ModelConfig__snapshot
[#0] PROGRESS:Eval -- Running for SigXsecOverSM = 1.8
=== Using the following for ModelConfig ===
Observables: RooArgSet:: = (obs_x_channel1,channelCat)
Parameters of Interest: RooArgSet:: = (SigXsecOverSM)
Nuisance Parameters: RooArgSet:: = (alpha_syst2,alpha_syst3,gamma_stat_channel1_bin_0,gamma_stat_channel1_bin_1)
Global Observables: RooArgSet:: = (nominalLumi,nom_alpha_syst1,nom_alpha_syst2,nom_alpha_syst3,nom_gamma_stat_channel1_bin_0,nom_gamma_stat_channel1_bin_1)
PDF: RooSimultaneous::simPdf[ indexCat=channelCat channel1=model_channel1 ] = 0.216225
Snapshot:
1) 0x9869e80 RooRealVar:: SigXsecOverSM = 1.8 L(0 - 3) "SigXsecOverSM"
=== Using the following for ModelConfig_with_poi_0 ===
Observables: RooArgSet:: = (obs_x_channel1,channelCat)
Parameters of Interest: RooArgSet:: = (SigXsecOverSM)
Nuisance Parameters: RooArgSet:: = (alpha_syst2,alpha_syst3,gamma_stat_channel1_bin_0,gamma_stat_channel1_bin_1)
Global Observables: RooArgSet:: = (nominalLumi,nom_alpha_syst1,nom_alpha_syst2,nom_alpha_syst3,nom_gamma_stat_channel1_bin_0,nom_gamma_stat_channel1_bin_1)
PDF: RooSimultaneous::simPdf[ indexCat=channelCat channel1=model_channel1 ] = 0.216225
Snapshot:
1) 0x9b73590 RooRealVar:: SigXsecOverSM = 0 L(0 - 3) "SigXsecOverSM"
[#0] PROGRESS:Generation -- Test Statistic on data: -2.05075
[#1] INFO:InputArguments -- Profiling conditional MLEs for Null.
[#1] INFO:InputArguments -- Using a ToyMCSampler. Now configuring for Null.
[#0] PROGRESS:Generation -- generated toys: 500 / 1000
[#1] INFO:InputArguments -- Profiling conditional MLEs for Alt.
[#1] INFO:InputArguments -- Using a ToyMCSampler. Now configuring for Alt.
[#0] PROGRESS:Eval -- P values for SigXsecOverSM = 1.8
CLs = 0.209052 +/- 0.0137241
CLb = 0.928 +/- 0.0115599
CLsplusb = 0.194 +/- 0.0125046
[#1] INFO:ObjectHandling -- RooWorkspace::saveSnapshot(combined) replacing previous snapshot with name ModelConfig__snapshot
[#0] PROGRESS:Eval -- Running for SigXsecOverSM = 2.4
=== Using the following for ModelConfig ===
Observables: RooArgSet:: = (obs_x_channel1,channelCat)
Parameters of Interest: RooArgSet:: = (SigXsecOverSM)
Nuisance Parameters: RooArgSet:: = (alpha_syst2,alpha_syst3,gamma_stat_channel1_bin_0,gamma_stat_channel1_bin_1)
Global Observables: RooArgSet:: = (nominalLumi,nom_alpha_syst1,nom_alpha_syst2,nom_alpha_syst3,nom_gamma_stat_channel1_bin_0,nom_gamma_stat_channel1_bin_1)
PDF: RooSimultaneous::simPdf[ indexCat=channelCat channel1=model_channel1 ] = 0.235304
Snapshot:
1) 0x99b9120 RooRealVar:: SigXsecOverSM = 2.4 L(0 - 3) "SigXsecOverSM"
=== Using the following for ModelConfig_with_poi_0 ===
Observables: RooArgSet:: = (obs_x_channel1,channelCat)
Parameters of Interest: RooArgSet:: = (SigXsecOverSM)
Nuisance Parameters: RooArgSet:: = (alpha_syst2,alpha_syst3,gamma_stat_channel1_bin_0,gamma_stat_channel1_bin_1)
Global Observables: RooArgSet:: = (nominalLumi,nom_alpha_syst1,nom_alpha_syst2,nom_alpha_syst3,nom_gamma_stat_channel1_bin_0,nom_gamma_stat_channel1_bin_1)
PDF: RooSimultaneous::simPdf[ indexCat=channelCat channel1=model_channel1 ] = 0.235304
Snapshot:
1) 0x99f6eb0 RooRealVar:: SigXsecOverSM = 0 L(0 - 3) "SigXsecOverSM"
[#0] PROGRESS:Generation -- Test Statistic on data: 0.0783908
[#1] INFO:InputArguments -- Profiling conditional MLEs for Null.
[#1] INFO:InputArguments -- Using a ToyMCSampler. Now configuring for Null.
[#0] PROGRESS:Generation -- generated toys: 500 / 1000
[#1] INFO:InputArguments -- Profiling conditional MLEs for Alt.
[#1] INFO:InputArguments -- Using a ToyMCSampler. Now configuring for Alt.
[#0] PROGRESS:Eval -- P values for SigXsecOverSM = 2.4
CLs = 0.0503282 +/- 0.00728062
CLb = 0.914 +/- 0.0125383
CLsplusb = 0.046 +/- 0.0066245
[#1] INFO:ObjectHandling -- RooWorkspace::saveSnapshot(combined) replacing previous snapshot with name ModelConfig__snapshot
[#0] PROGRESS:Eval -- Running for SigXsecOverSM = 3
=== Using the following for ModelConfig ===
Observables: RooArgSet:: = (obs_x_channel1,channelCat)
Parameters of Interest: RooArgSet:: = (SigXsecOverSM)
Nuisance Parameters: RooArgSet:: = (alpha_syst2,alpha_syst3,gamma_stat_channel1_bin_0,gamma_stat_channel1_bin_1)
Global Observables: RooArgSet:: = (nominalLumi,nom_alpha_syst1,nom_alpha_syst2,nom_alpha_syst3,nom_gamma_stat_channel1_bin_0,nom_gamma_stat_channel1_bin_1)
PDF: RooSimultaneous::simPdf[ indexCat=channelCat channel1=model_channel1 ] = 0.254383
Snapshot:
1) 0x9b6c650 RooRealVar:: SigXsecOverSM = 3 L(0 - 3) "SigXsecOverSM"
=== Using the following for ModelConfig_with_poi_0 ===
Observables: RooArgSet:: = (obs_x_channel1,channelCat)
Parameters of Interest: RooArgSet:: = (SigXsecOverSM)
Nuisance Parameters: RooArgSet:: = (alpha_syst2,alpha_syst3,gamma_stat_channel1_bin_0,gamma_stat_channel1_bin_1)
Global Observables: RooArgSet:: = (nominalLumi,nom_alpha_syst1,nom_alpha_syst2,nom_alpha_syst3,nom_gamma_stat_channel1_bin_0,nom_gamma_stat_channel1_bin_1)
PDF: RooSimultaneous::simPdf[ indexCat=channelCat channel1=model_channel1 ] = 0.254383
Snapshot:
1) 0x9b501b0 RooRealVar:: SigXsecOverSM = 0 L(0 - 3) "SigXsecOverSM"
[#0] PROGRESS:Generation -- Test Statistic on data: 3.27476
[#1] INFO:InputArguments -- Profiling conditional MLEs for Null.
[#1] INFO:InputArguments -- Using a ToyMCSampler. Now configuring for Null.
[#0] PROGRESS:Generation -- generated toys: 500 / 1000
[#1] INFO:InputArguments -- Profiling conditional MLEs for Alt.
[#1] INFO:InputArguments -- Using a ToyMCSampler. Now configuring for Alt.
[#0] PROGRESS:Eval -- P values for SigXsecOverSM = 3
CLs = 0.00534188 +/- 0.0023838
CLb = 0.936 +/- 0.0109457
CLsplusb = 0.005 +/- 0.00223047
Time to perform limit scan
Real time 0:00:04, CP time 4.490
The computed upper limit is: 2.40438 +/- 0.0566658
Expected upper limits, using the B (alternate) model :
expected limit (median) 1.61927
expected limit (-1 sig) 1.09318
expected limit (+1 sig) 2.24198
expected limit (-2 sig) 0.787047
expected limit (+2 sig) 2.86153
[#0] WARNING:Plotting -- Could not determine xmin and xmax of sampling distribution that was given to plot.
[#0] WARNING:Plotting -- Could not determine xmin and xmax of sampling distribution that was given to plot.
#include <cassert>
using std::cout, std::endl;
bool optimize = true;
bool generateBinned = false;
false;
std::string minimizerType =
"";
};
public:
void SetParameter(
const char *
name,
const char *
value);
void SetParameter(
const char *
name,
bool value);
void SetParameter(
const char *
name,
int value);
void SetParameter(
const char *
name,
double value);
private:
std::string
};
}
RooStats::HypoTestInvTool::HypoTestInvTool()
{
}
void RooStats::HypoTestInvTool::SetParameter(
const char *
name,
bool value)
{
if (
s_name.find(
"PlotHypoTestResult") != std::string::npos)
if (
s_name.find(
"WriteResult") != std::string::npos)
if (
s_name.find(
"Optimize") != std::string::npos)
if (
s_name.find(
"UseVectorStore") != std::string::npos)
if (
s_name.find(
"GenerateBinned") != std::string::npos)
if (
s_name.find(
"UseProof") != std::string::npos)
if (
s_name.find(
"EnableDetailedOutput") != std::string::npos)
if (
s_name.find(
"Rebuild") != std::string::npos)
if (
s_name.find(
"ReuseAltToys") != std::string::npos)
return;
}
void RooStats::HypoTestInvTool::SetParameter(
const char *
name,
int value)
{
if (
s_name.find(
"NWorkers") != std::string::npos)
if (
s_name.find(
"NToyToRebuild") != std::string::npos)
if (
s_name.find(
"RebuildParamValues") != std::string::npos)
if (
s_name.find(
"PrintLevel") != std::string::npos)
if (
s_name.find(
"InitialFit") != std::string::npos)
if (
s_name.find(
"RandomSeed") != std::string::npos)
if (
s_name.find(
"AsimovBins") != std::string::npos)
return;
}
void RooStats::HypoTestInvTool::SetParameter(
const char *
name,
double value)
{
if (
s_name.find(
"NToysRatio") != std::string::npos)
if (
s_name.find(
"MaxPOI") != std::string::npos)
return;
}
void RooStats::HypoTestInvTool::SetParameter(
const char *
name,
const char *
value)
{
if (
s_name.find(
"MassValue") != std::string::npos)
if (
s_name.find(
"MinimizerType") != std::string::npos)
if (
s_name.find(
"ResultFileName") != std::string::npos)
return;
}
{
filename =
"results/example_combined_GaussExample_model.root";
cout << "will run standard hist2workspace example" << endl;
gROOT->ProcessLine(
".! prepareHistFactory .");
gROOT->ProcessLine(
".! hist2workspace config/example.xml");
cout << "\n\n---------------------" << endl;
cout << "Done creating example input" << endl;
cout << "---------------------\n\n" << endl;
}
} else
if (!file) {
cout <<
"StandardRooStatsDemoMacro: Input file " <<
filename <<
" is not found" << endl;
return;
}
calc.SetParameter(
"PlotHypoTestResult",
optHTInv.plotHypoTestResult);
calc.SetParameter(
"UseVectorStore",
optHTInv.useVectorStore);
calc.SetParameter(
"GenerateBinned",
optHTInv.generateBinned);
calc.SetParameter(
"EnableDetailedOutput",
optHTInv.enableDetailedOutput);
calc.SetParameter(
"RebuildParamValues",
optHTInv.rebuildParamValues);
calc.SetParameter(
"MinimizerType",
optHTInv.minimizerType.c_str());
calc.SetParameter(
"ResultFileName",
optHTInv.resultFileName);
std::cout <<
w <<
"\t" <<
filename << std::endl;
std::cerr << "Error running the HypoTestInverter - Exit " << std::endl;
return;
}
} else {
std::cout <<
"Reading an HypoTestInverterResult with name " <<
wsName <<
" from file " <<
filename << std::endl;
std::cerr <<
"File " <<
filename <<
" does not contain a workspace or an HypoTestInverterResult - Exit " << std::endl;
return;
}
}
return;
}
{
#if defined ROOT_SVN_VERSION && ROOT_SVN_VERSION >= 44126
llError =
r->LowerLimitEstimatedError();
}
#else
llError =
r->LowerLimitEstimatedError();
#endif
double ulError =
r->UpperLimitEstimatedError();
std::cout <<
"The computed lower limit is: " <<
lowerLimit <<
" +/- " <<
llError << std::endl;
std::cout <<
"The computed upper limit is: " <<
upperLimit <<
" +/- " <<
ulError << std::endl;
std::cout << "Expected upper limits, using the B (alternate) model : " << std::endl;
std::cout <<
" expected limit (median) " <<
r->GetExpectedUpperLimit(0) << std::endl;
std::cout <<
" expected limit (-1 sig) " <<
r->GetExpectedUpperLimit(-1) << std::endl;
std::cout <<
" expected limit (+1 sig) " <<
r->GetExpectedUpperLimit(1) << std::endl;
std::cout <<
" expected limit (-2 sig) " <<
r->GetExpectedUpperLimit(-2) << std::endl;
std::cout <<
" expected limit (+2 sig) " <<
r->GetExpectedUpperLimit(2) << std::endl;
Info(
"StandardHypoTestInvDemo",
"detailed output will be written in output result file");
}
}
name.Replace(0,
name.Last(
'/') + 1,
"");
}
}
Info(
"StandardHypoTestInvDemo",
"HypoTestInverterResult has been written in the file %s",
mResultFileName.Data());
}
std::string typeName = "";
typeName = "Frequentist";
typeName = "Hybrid";
typeName = "Asymptotic";
}
}
}
}
}
{
std::cout <<
"Running HypoTestInverter on the workspace " <<
w->GetName() << std::endl;
return nullptr;
} else
std::cout <<
"Using data set " <<
dataName << std::endl;
data->convertToVectorStore();
}
return nullptr;
}
return nullptr;
}
if (!
sbModel->GetParametersOfInterest()) {
return nullptr;
}
return nullptr;
}
Info(
"StandardHypoTestInvDemo",
"Model %s has no snapshot - make one using model poi",
modelSBName);
}
std::cout << "StandardHypoTestInvDemo"
<< " - Switch off all systematics by setting them constant to their initial values" << std::endl;
}
}
}
Info(
"StandardHypoTestInvDemo",
"The background model %s does not exist",
modelBName);
Info(
"StandardHypoTestInvDemo",
"Copy it from ModelConfig %s and set POI to zero",
modelSBName);
if (!var)
return nullptr;
} else {
Info(
"StandardHypoTestInvDemo",
"Model %s has no snapshot - make one using model poi and 0 values ",
if (var) {
} else {
return nullptr;
}
}
}
Warning(
"StandardHypoTestInvDemo",
"Model %s has nuisance parameters but no global observables associated",
"\tThe effect of the nuisance parameters will not be treated correctly ");
}
}
}
std::cout <<
"StandardHypoTestInvDemo : POI initial value: " << poi->
GetName() <<
" = " << poi->
getVal()
<< std::endl;
doFit = false;
doFit = false;
else
Info(
"StandardHypoTestInvDemo",
"Using %s as minimizer for computing the test statistic",
if (doFit) {
Info(
"StandardHypoTestInvDemo",
" Doing a first fit to the observed data ");
if (
sbModel->GetNuisanceParameters())
"Fit to the model failed - try with strategy 1 and perform first an Hesse computation");
}
Warning(
"StandardHypoTestInvDemo",
" Fit still failed - continue anyway.....");
std::cout <<
"StandardHypoTestInvDemo - Best Fit value : " << poi->
GetName() <<
" = " <<
poihat <<
" +/- " std::cout << "Time for fitting : ";
std::cout <<
"StandardHypoTestInvo: snapshot of S+B Model " <<
sbModel->GetName()
<< " is set to the best fit value" << std::endl;
}
if (!doFit)
Info(
"StandardHypoTestInvDemo",
"Using LEP test statistic - an initial fit is not done and the TS will use "
"the nuisances at the model value");
else
Info(
"StandardHypoTestInvDemo",
"Using LEP test statistic - an initial fit has been done and the TS will use "
"the nuisances at the best fit value");
}
if (
sbModel->GetNuisanceParameters())
if (
bModel->GetNuisanceParameters())
slrts.EnableDetailedOutput();
ropl.SetSubtractMLE(
false);
ropl.SetSubtractMLE(
true);
ropl.EnableDetailedOutput();
profll.EnableDetailedOutput();
}
true);
else {
Error(
"StandardHypoTestInvDemo",
"Invalid - calculator type = %d supported values are only :\n\t\t\t 0 "
"(Frequentist) , 1 (Hybrid) , 2 (Asymptotic) ",
return nullptr;
}
Error(
"StandardHypoTestInvDemo",
"Invalid - test statistic type = %d supported values are only :\n\t\t\t 0 (SLR) "
", 1 (Tevatron) , 2 (PLR), 3 (PLR1), 4(MLE)",
return nullptr;
}
if (
sbModel->GetPdf()->canBeExtended()) {
Warning(
"StandardHypoTestInvDemo",
"Pdf is extended: but number counting flag is set: ignore it ");
} else {
int nEvents =
data->numEntries();
Info(
"StandardHypoTestInvDemo",
"Pdf is not extended: number of events to generate taken from observed data set is %d", nEvents);
toymcs->SetNEventsPerToy(nEvents);
} else {
Info(
"StandardHypoTestInvDemo",
"using a number counting pdf");
}
}
Info(
"StandardHypoTestInvDemo",
"Data set is weighted, nentries = %d and sum of weights = %8.1f but toy "
"generation is unbinned - it would be faster to set mGenerateBinned to true\n",
data->numEntries(),
data->sumEntries());
}
Warning(
"StandardHypoTestInvDemo",
"generate binned is activated but the number of observable is %d. Too much "
"memory could be needed for allocating all the bins",
sbModel->GetObservables()->getSize());
}
}
}
if (
bModel->GetNuisanceParameters() ||
sbModel->GetNuisanceParameters()) {
toymcs->SetUseMultiGen(
false);
ToyMCSampler::SetAlwaysUseMultiGen(false);
Info(
"StandardHypoTestInvDemo",
"No nuisance pdf given for the HybridCalculator - try to deduce pdf from the model");
else
}
Info(
"StandardHypoTestInvDemo",
"No nuisance pdf given - try to use %s that is defined as a prior pdf in the B model",
} else {
Error(
"StandardHypoTestInvDemo",
"Cannot run Hybrid calculator because no prior on the nuisance "
"parameter is specified or can be derived");
return nullptr;
}
}
Info(
"StandardHypoTestInvDemo",
"Using as nuisance Pdf ... ");
(
bModel->GetNuisanceParameters()) ?
bModel->GetNuisanceParameters() :
sbModel->GetNuisanceParameters();
if (
np->getSize() == 0) {
"Prior nuisance does not depend on nuisance parameters. They will be smeared in their full range");
}
}
"Only the PL test statistic can be used with AsymptoticCalculator - use by default a two-sided PL");
}
}
}
std::cout <<
"Doing a fixed scan in interval : " <<
poimin <<
" , " <<
poimax << std::endl;
} else {
std::cout <<
"Doing an automatic scan in interval : " << poi->
getMin() <<
" , " << poi->
getMax() << std::endl;
}
std::cout << "Time to perform limit scan \n";
std::cout << "\n***************************************************************\n";
std::cout << "Rebuild the upper limit distribution by re-generating new set of pseudo-experiment and re-compute "
"for each of them a new upper limit\n\n";
}
if (
sbModel->GetNuisanceParameters())
std::cout << "rebuild using fitted parameter value for B-model snapshot" << std::endl;
}
}
std::cout << "StandardHypoTestInvDemo: Initial parameters used for rebuilding: ";
calc.SetCloseProof(
true);
std::cout << "Time to rebuild distributions " << std::endl;
std::cout << "Expected limits after rebuild distribution " << std::endl;
std::cout <<
"expected upper limit (median of limit distribution) " <<
limDist->InverseCDF(0.5) << std::endl;
std::cout << "expected -1 sig limit (0.16% quantile of limit dist) "
std::cout << "expected +1 sig limit (0.84% quantile of limit dist) "
std::cout << "expected -2 sig limit (.025% quantile of limit dist) "
std::cout << "expected +2 sig limit (.975% quantile of limit dist) "
new TCanvas(
"limPlot",
"Upper Limit Distribution");
} else
std::cout << "ERROR : failed to re-build distributions " << std::endl;
}
}
{
}
#ifdef USE_AS_MAIN
{
}
#endif
void Info(const char *location, const char *msgfmt,...)
Use this function for informational messages.
void Error(const char *location, const char *msgfmt,...)
Use this function in case an error occurred.
void Warning(const char *location, const char *msgfmt,...)
Use this function in warning situations.
winID h TVirtualViewer3D TVirtualGLPainter char TVirtualGLPainter plot
Option_t Option_t TPoint TPoint const char GetTextMagnitude GetFillStyle GetLineColor GetLineWidth GetMarkerStyle GetTextAlign GetTextColor GetTextSize void char Point_t Rectangle_t WindowAttributes_t Float_t Float_t Float_t Int_t Int_t UInt_t UInt_t Rectangle_t Int_t Int_t Window_t TString Int_t GCValues_t GetPrimarySelectionOwner GetDisplay GetScreen GetColormap GetNativeEvent const char const char dpyName wid window const char font_name cursor keysym reg const char only_if_exist regb h Point_t winding char text const char depth char const char Int_t count const char ColorStruct_t color const char filename
Option_t Option_t TPoint TPoint const char GetTextMagnitude GetFillStyle GetLineColor GetLineWidth GetMarkerStyle GetTextAlign GetTextColor GetTextSize void char Point_t Rectangle_t WindowAttributes_t Float_t Float_t Float_t Int_t Int_t UInt_t UInt_t Rectangle_t Int_t Int_t Window_t TString Int_t GCValues_t GetPrimarySelectionOwner GetDisplay GetScreen GetColormap GetNativeEvent const char const char dpyName wid window const char font_name cursor keysym reg const char only_if_exist regb h Point_t np
Option_t Option_t TPoint TPoint const char GetTextMagnitude GetFillStyle GetLineColor GetLineWidth GetMarkerStyle GetTextAlign GetTextColor GetTextSize void char Point_t Rectangle_t WindowAttributes_t Float_t r
Option_t Option_t TPoint TPoint const char GetTextMagnitude GetFillStyle GetLineColor GetLineWidth GetMarkerStyle GetTextAlign GetTextColor GetTextSize void value
R__EXTERN TSystem * gSystem
static void SetDefaultMinimizer(const char *type, const char *algo=nullptr)
Set the default Minimizer type and corresponding algorithms.
static void SetDefaultStrategy(int strat)
Set the default strategy.
static const std::string & DefaultMinimizerType()
Abstract base class for binned and unbinned datasets.
static void setDefaultStorageType(StorageType s)
Abstract interface for all probability density functions.
virtual double getMax(const char *name=nullptr) const
Get maximum of currently defined range.
virtual double getMin(const char *name=nullptr) const
Get minimum of currently defined range.
double getVal(const RooArgSet *normalisationSet=nullptr) const
Evaluate object.
RooArgSet is a container object that can hold multiple RooAbsArg objects.
static RooMsgService & instance()
Return reference to singleton instance.
static TRandom * randomGenerator()
Return a pointer to a singleton random-number generator implementation.
Variable that can be changed from the outside.
void setVal(double value) override
Set value of variable to 'value'.
void setMax(const char *name, double value)
Set maximum of name range to given value.
Hypothesis Test Calculator based on the asymptotic formulae for the profile likelihood ratio.
Does a frequentist hypothesis test.
Same purpose as HybridCalculatorOriginal, but different implementation.
Common base class for the Hypothesis Test Calculators.
Class to plot a HypoTestInverterResult, the output of the HypoTestInverter calculator.
HypoTestInverterResult class holds the array of hypothesis test results and compute a confidence inte...
A class for performing a hypothesis test inversion by scanning the hypothesis test results of a HypoT...
MaxLikelihoodEstimateTestStat: TestStatistic that returns maximum likelihood estimate of a specified ...
ModelConfig is a simple class that holds configuration information specifying how a model should be u...
NumEventsTestStat is a simple implementation of the TestStatistic interface used for simple number co...
ProfileLikelihoodTestStat is an implementation of the TestStatistic interface that calculates the pro...
Holds configuration options for proof and proof-lite.
TestStatistic that returns the ratio of profiled likelihoods.
This class provides simple and straightforward utilities to plot SamplingDistribution objects.
This class simply holds a sampling distribution of some test statistic.
TestStatistic class that returns -log(L[null] / L[alt]) where L is the likelihood.
TestStatistic is an interface class to provide a facility for construction test statistics distributi...
ToyMCSampler is an implementation of the TestStatSampler interface.
Persistable container for RooFit projects.
TObject * Get(const char *namecycle) override
Return pointer to object identified by namecycle.
A ROOT file is an on-disk file, usually with extension .root, that stores objects in a file-system-li...
void ls(Option_t *option="") const override
List file contents.
static TFile * Open(const char *name, Option_t *option="", const char *ftitle="", Int_t compress=ROOT::RCompressionSetting::EDefaults::kUseCompiledDefault, Int_t netopt=0)
Create / open a file.
const char * GetName() const override
Returns name of object.
Mother of all ROOT objects.
static TString Format(const char *fmt,...)
Static method which formats a string using a printf style format descriptor and return a TString.
virtual Bool_t AccessPathName(const char *path, EAccessMode mode=kFileExists)
Returns FALSE if one can access a file using the specified access mode.
RooCmdArg InitialHesse(bool flag=true)
RooCmdArg Offset(std::string const &mode)
RooCmdArg Constrain(const RooArgSet ¶ms)
RooCmdArg Minimizer(const char *type, const char *alg=nullptr)
RooCmdArg Hesse(bool flag=true)
RooCmdArg Strategy(Int_t code)
RooCmdArg Save(bool flag=true)
RooCmdArg PrintLevel(Int_t code)
double normal_cdf(double x, double sigma=1, double x0=0)
Cumulative distribution function of the normal (Gaussian) distribution (lower tail).
The namespace RooFit contains mostly switches that change the behaviour of functions of PDFs (or othe...
Namespace for the RooStats classes.
bool SetAllConstant(const RooAbsCollection &coll, bool constant=true)
utility function to set all variable constant in a collection (from G.
void RemoveConstantParameters(RooArgSet *set)
RooAbsPdf * MakeNuisancePdf(RooAbsPdf &pdf, const RooArgSet &observables, const char *name)
extract constraint terms from pdf
void UseNLLOffset(bool on)
function to set a global flag in RooStats to use NLL offset when performing nll computations Note tha...
bool IsNLLOffset()
function returning if the flag to check if the flag to use NLLOffset is set
void PrintListContent(const RooArgList &l, std::ostream &os=std::cout)
useful function to print in one line the content of a set with their values
Double_t Sqrt(Double_t x)
Returns the square root of x.
Int_t CeilNint(Double_t x)
Returns the nearest integer of TMath::Ceil(x).