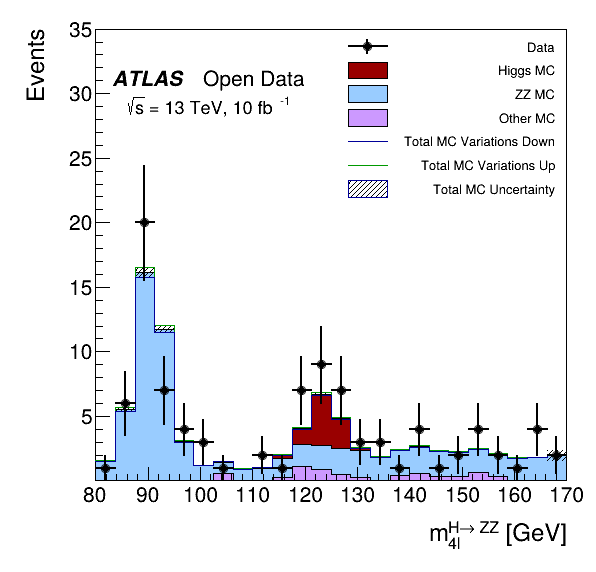
This tutorial is the Higgs to four lepton analysis from the ATLAS Open Data release in 2020 (http://opendata.atlas.cern/release/2020/documentation/). The data was taken with the ATLAS detector during 2016 at a center-of-mass energy of 13 TeV. The decay of the Standard Model Higgs boson to two Z bosons and subsequently to four leptons is called the "golden channel". The selection leads to a narrow invariant mass peak on top a relatively smooth and small background, revealing the Higgs at 125 GeV. Systematic errors for the MC scale factors are computed and the Vary function of RDataFrame is used for plotting. The analysis is translated to an RDataFrame workflow processing about 300 MB of simulated events and data.
import ROOT
import os
"""
float scale(unsigned int slot, const ROOT::RDF::RSampleInfo &id){
return id.Contains("mc_363490.llll.4lep.root") ? 1.3f : 1.0f;
}
"""
)
"""
using ROOT::RVecF;
using ROOT::RVecI;
bool GoodElectronsAndMuons(const RVecI &type, const RVecF &pt, const RVecF &eta, const RVecF &phi, const RVecF &e, const RVecF &trackd0pv, const RVecF &tracksigd0pv, const RVecF &z0)
{
for (size_t i = 0; i < type.size(); i++) {
ROOT::Math::PtEtaPhiEVector p(0.001*pt[i], eta[i], phi[i], 0.001*e[i]);
if (type[i] == 11) {
if (pt[i] < 7000 || abs(eta[i]) > 2.47 || abs(trackd0pv[i] / tracksigd0pv[i]) > 5 || abs(z0[i] * sin(p.Theta())) > 0.5) return false;
} else {
if (abs(trackd0pv[i] / tracksigd0pv[i]) > 5 || abs(z0[i] * sin(p.Theta())) > 0.5) return false;
}
}
return true;
}
"""
)
df = (
"good_lep",
"abs(lep_eta) < 2.5 && lep_pt > 5000 && lep_ptcone30 / lep_pt < 0.3 && lep_etcone20 / lep_pt < 0.3",
)
.Filter("Sum(good_lep) == 4")
.Filter("Sum(lep_charge[good_lep]) == 0")
.Define("goodlep_sumtypes", "Sum(lep_type[good_lep])")
.Filter("goodlep_sumtypes == 44 || goodlep_sumtypes == 52 || goodlep_sumtypes == 48")
)
"GoodElectronsAndMuons(lep_type[good_lep], lep_pt[good_lep], lep_eta[good_lep], lep_phi[good_lep], lep_E[good_lep], lep_trackd0pvunbiased[good_lep], lep_tracksigd0pvunbiased[good_lep], lep_z0[good_lep])"
)
df = (
.Define("goodlep_eta", "lep_eta[good_lep]")
.Define("goodlep_phi", "lep_phi[good_lep]")
.Define("goodlep_E", "lep_E[good_lep]")
.Define("goodlep_type", "lep_type[good_lep]")
)
df =
df.Filter(
"goodlep_pt[0] > 25000 && goodlep_pt[1] > 15000 && goodlep_pt[2] > 10000")
"""
double weights(float scaleFactor_1, float scaleFactor_2, float scaleFactor_3, float scaleFactor_4, float scale, float mcWeight, double xsecs, double sumws, double lumi)
{
return scaleFactor_1 * scaleFactor_2 * scaleFactor_3 * scaleFactor_4 * scale * mcWeight * xsecs / sumws * lumi;
}
"""
)
"weight",
"double x; return isMC ? weights(scaleFactor_ELE, scaleFactor_MUON, scaleFactor_LepTRIGGER, scaleFactor_PILEUP, scale, mcWeight, xsecs, sumws, lumi) : 1.;",
)
"""
float ComputeInvariantMass(RVecF pt, RVecF eta, RVecF phi, RVecF e)
{
ROOT::Math::PtEtaPhiEVector p1{pt[0], eta[0], phi[0], e[0]};
ROOT::Math::PtEtaPhiEVector p2{pt[1], eta[1], phi[1], e[1]};
ROOT::Math::PtEtaPhiEVector p3{pt[2], eta[2], phi[2], e[2]};
ROOT::Math::PtEtaPhiEVector p4{pt[3], eta[3], phi[3], e[3]};
return 0.001 * (p1 + p2 + p3 + p4).M();
}
"""
)
df =
df.Define(
"m4l",
"ComputeInvariantMass(goodlep_pt, goodlep_eta, goodlep_phi, goodlep_E)")
histos = []
for sample_category in ["data", "higgs", "zz", "other"]:
df.Filter(f
'sample_category == "{sample_category}"').Histo1D(
"m4l",
"weight",
)
)
"""
using namespace ROOT::VecOps;
class VaryHelper
{
const std::vector<double> x{5.50e3, 5.52e3, 12.54e3, 17.43e3, 22.40e3, 27.48e3, 30e3, 10000e3};
const std::vector<double> y{0.06628, 0.06395, 0.06396, 0.03372, 0.02441, 0.01403, 0, 0};
TGraph graph;
public:
VaryHelper() : graph(x.size(), x.data(), y.data()) {}
RVec<double> operator()(const double &w, const RVecF &pt, const RVec<unsigned int> &type)
{
const auto v = Mean(Map(pt[type == 11], [this](auto p)
{return this->graph.Eval(p); })
);
return RVec<double>{(1 + v) * w, (1 - v) * w};
}
};
VaryHelper variationsFactory;
"""
)
df_variations_mc = (
.Vary("weight", "variationsFactory(weight, goodlep_pt, goodlep_type)", ["up", "down"])
)
for i
in range(0, histos_mc[
"nominal"].GetXaxis().GetNbins()):
(
histos_mc["nominal"].SetBinError(
i, (histos_mc["weight:up"].GetBinContent(i) - histos_mc["nominal"].GetBinContent(i))
)
)
h_data = histos[0].GetValue().Clone()
h_higgs = histos[1].GetValue().Clone()
h_zz = histos[2].GetValue().Clone()
h_other = histos[3].GetValue().Clone()
h_nominal = histos_mc["nominal"].DrawClone("E2 same")
h_weight_up = histos_mc["weight:up"].DrawClone("HIST SAME")
h_weight_down = histos_mc["weight:down"].DrawClone("HIST SAME")
c1.SaveAs(
"df106_HiggsToFourLeptons_python.png")
print("Saved figure to df106_HiggsToFourLeptons_python.png")
ROOT::Detail::TRangeCast< T, true > TRangeDynCast
TRangeDynCast is an adapter class that allows the typed iteration through a TCollection.
Option_t Option_t SetFillStyle
Option_t Option_t SetLineColor
Option_t Option_t SetFillColor
A struct which stores the parameters of a TH1D.