This tutorial is the analysis of single top production adapted from the ATLAS Open Data release in 2020 (http://opendata.atlas.cern/release/2020/documentation/). The data was recorded with the ATLAS detector during 2016 at a center-of-mass energy of 13 TeV. Top quarks with a mass of about 172 GeV are mostly produced in pairs but also appear alone, dominantly from the decays of a W boson in association with a light jet.
The analysis is translated to a RDataFrame workflow processing up to 60 GB of simulated events and data. By default the analysis runs on a preskimmed dataset to reduce the runtime. The full dataset can be used with the –full-dataset argument and you can also run only on a fraction of the original dataset using the argument –lumi-scale.
import ROOT
import json
import argparse
import os
parser = argparse.ArgumentParser()
parser.add_argument("--lumi-scale", type=float, default=0.05,
help="Run only on a fraction of the total available 10 fb^-1 (only usable together with --full-dataset)")
parser.add_argument("--full-dataset", action="store_true", default=False,
help="Use the full dataset (use --lumi-scale to run only on a fraction of it)")
parser.add_argument("-b", action="store_true", default=False, help="Use ROOT batch mode")
parser.add_argument("-t", action="store_true", default=False, help="Use implicit multi threading (for the full dataset only possible with --lumi-scale 1.0)")
args = parser.parse_args()
if args.b: ROOT.gROOT.SetBatch(True)
if not args.full_dataset: lumi_scale = 0.05
else: lumi_scale = args.lumi_scale
lumi = 10064.0
print('Run on data corresponding to {:.1f} fb^-1 ...'.format(lumi * lumi_scale / 1000.0))
if args.full_dataset: dataset_path = "root://eospublic.cern.ch//eos/opendata/atlas/OutreachDatasets/2020-01-22"
else: dataset_path = "root://eospublic.cern.ch//eos/root-eos/reduced_atlas_opendata/singletop"
files = json.load(open(os.path.join(os.path.dirname(os.path.abspath(__file__)), "df107_SingleTopAnalysis.json")))
processes = files.keys()
df = {}
xsecs = {}
sumws = {}
samples = []
for p in processes:
for d in files[p]:
folder = d[0]
sample = d[1]
xsecs[sample] = d[2]
sumws[sample] = d[3]
num_events = d[4]
samples.append(sample)
df[sample] =
ROOT.RDataFrame(
"mini",
"{}/1lep/{}/{}.1lep.root".format(dataset_path, folder, sample))
if args.full_dataset and lumi_scale < 1.0:
df[sample] = df[sample].
Range(
int(num_events * lumi_scale))
ROOT.gInterpreter.Declare("""
using cRVecF = const ROOT::RVecF &;
using cRVecI = const ROOT::RVecI &;
int FindGoodLepton(cRVecI goodlep, cRVecI type, cRVecF lep_pt, cRVecF lep_eta, cRVecF lep_phi, cRVecF lep_e, cRVecF trackd0pv, cRVecF tracksigd0pv, cRVecF z0)
{
int idx = -1; // Return -1 if no good lepton is found.
for(auto i = 0; i < type.size(); i++) {
if(!goodlep[i]) continue;
if (type[i] == 11 && abs(lep_eta[i]) < 2.47 && (abs(lep_eta[i]) < 1.37 || abs(lep_eta[i]) > 1.52) && abs(trackd0pv[i] / tracksigd0pv[i]) < 5) {
const ROOT::Math::PtEtaPhiEVector p(lep_pt[i], lep_eta[i], lep_phi[i], lep_e[i]);
if (abs(z0[i] * sin(p.Theta())) < 0.5) {
if (idx == -1) idx = i;
else return -1; // Accept only events with exactly one good lepton
}
}
if (type[i] == 13 && abs(lep_eta[i]) < 2.5 && abs(trackd0pv[i] / tracksigd0pv[i]) < 3) {
const ROOT::Math::PtEtaPhiEVector p(lep_pt[i], lep_eta[i], lep_phi[i], lep_e[i]);
if (abs(z0[i] * sin(p.Theta())) < 0.5) {
if (idx == -1) idx = i;
else return -1; // Accept only events with exactly one good lepton
}
}
}
return idx;
}
""")
for s in samples:
df[s] = df[s].Filter("trigE || trigM")\
.Filter("met_et > 30000")
df[s] = df[s].Define("goodlep", "lep_isTightID && lep_pt > 35000 && lep_ptcone30 / lep_pt < 0.1 && lep_etcone20 / lep_pt < 0.1")\
.Filter("ROOT::VecOps::Sum(goodlep) > 0")
df[s] = df[s].Define("idx_lep", "FindGoodLepton(goodlep, lep_type, lep_pt, lep_eta, lep_phi, lep_E, lep_trackd0pvunbiased, lep_tracksigd0pvunbiased, lep_z0)")\
.Filter("idx_lep != -1")
df[s] = df[s].Define("mtw", "sqrt(2 * lep_pt[idx_lep] * met_et * (1 - cos(lep_phi[idx_lep] - met_phi)))")\
.Filter("mtw > 60000")
df[s] = df[s].Filter("ROOT::VecOps::Sum(jet_pt > 30000 && abs(jet_eta) < 2.5) > 0")
df[s] = df[s].Define("goodjet", "jet_pt > 60000 || abs(jet_eta) > 2.4 || jet_jvt > 0.59")\
.Filter("ROOT::VecOps::Sum(goodjet) == 2")\
.Define("goodbjet", "goodjet && jet_MV2c10 > 0.8244273")\
.Filter("ROOT::VecOps::Sum(goodbjet) == 1")\
.Define("idx_tagged", "ROOT::VecOps::ArgMax(goodjet && goodbjet)")\
.Define("idx_untagged", "ROOT::VecOps::ArgMax(goodjet && !goodbjet)")
df[s] = df[s].Filter("abs(jet_eta[idx_untagged]) > 1.5 && abs(jet_eta[idx_tagged] - jet_eta[idx_untagged]) > 1.5")\
.Filter("lep_pt[idx_lep] + jet_pt[idx_tagged] + jet_pt[idx_untagged] + met_et > 195000")
for s in samples:
if "data" in s:
df[s] = df[s].Define("weight", "1.0")
else:
if "single" in s: stop_norm = "mcWeight / abs(mcWeight)"
else: stop_norm = "mcWeight"
df[s] = df[s].Define("weight", "scaleFactor_ELE * scaleFactor_MUON * scaleFactor_LepTRIGGER * scaleFactor_PILEUP * scaleFactor_BTAG * {} * {} / {} * {}".format(stop_norm, xsecs[s], sumws[s], lumi))
ROOT.gInterpreter.Declare("""
float ComputeTopMass(float lep_pt, float lep_eta, float lep_phi, float lep_e, float jet_pt, float jet_eta, float jet_phi, float jet_e, float met_et, float met_phi)
{
const ROOT::Math::PtEtaPhiEVector lep(lep_pt / 1000.0, lep_eta, lep_phi, lep_e / 1000.0);
const ROOT::Math::PtEtaPhiEVector met(met_et / 1000.0, 0, met_phi, met_et / 1000.0);
const ROOT::Math::PtEtaPhiEVector bjet(jet_pt / 1000.0, jet_eta, jet_phi, jet_e / 1000.0);
// Please note that we treat here the missing transverse energy as the neutrino, even though the z component is missing!
return (lep + met + bjet).M();
}
""")
histos = {}
for s in samples:
df[s] = df[s].Define("top_mass", "ComputeTopMass(lep_pt[idx_lep], lep_eta[idx_lep], lep_phi[idx_lep], lep_E[idx_lep], jet_pt[idx_tagged], jet_eta[idx_tagged], jet_phi[idx_tagged], jet_E[idx_tagged], met_et, met_phi)")
histos[s] = df[s].Histo1D(
ROOT.RDF.TH1DModel(
"top_mass",
"", 10, 100, 400),
"top_mass",
"weight")
def merge_histos(label):
h = None
for i, d in enumerate(files[label]):
t = histos[d[1]].GetValue()
if i == 0: h = t.Clone()
else: h.Add(t)
h.SetNameTitle(label, label)
return h
data = merge_histos("data")
twtb = merge_histos("twtb")
singletop = merge_histos("singletop")
wjets = merge_histos("wjets")
ROOT.gROOT.SetStyle("ATLAS")
c = ROOT.TCanvas("c", "", 600, 600)
pad = ROOT.TPad("upper_pad", "", 0, 0, 1, 1)
pad.SetTickx(False)
pad.SetTicky(False)
pad.Draw()
pad.cd()
stack = ROOT.THStack()
wjets.Scale(1.1)
for h, color in zip(
[wjets, twtb, singletop],
[(222, 90, 106), (155, 152, 204), (208, 240, 193)]):
h.SetLineWidth(1)
h.SetLineColor(1)
h.SetFillColor(ROOT.TColor.GetColor(*color))
stack.Add(h)
stack.Draw("HIST")
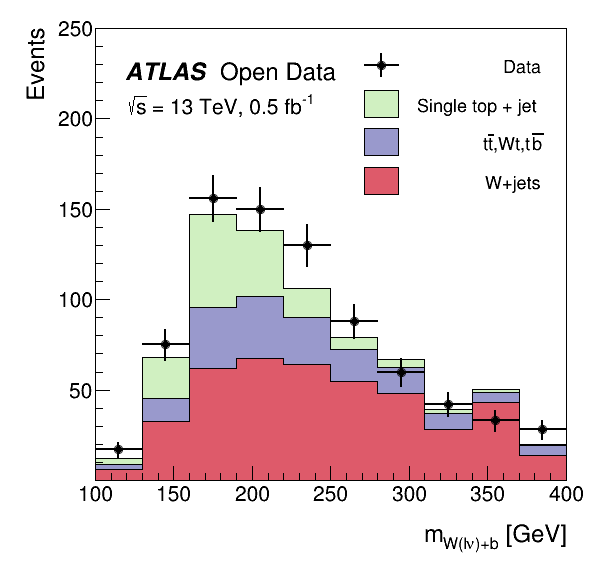
stack.GetXaxis().SetTitle("m_{W(l#nu)+b} [GeV]")
stack.GetYaxis().SetTitle("Events")
stack.GetYaxis().SetLabelSize(0.04)
stack.GetYaxis().SetTitleSize(0.045)
stack.GetXaxis().SetLabelSize(0.04)
stack.GetXaxis().SetTitleSize(0.045)
stack.SetMinimum(0)
stack.SetMaximum(5000 * lumi_scale)
stack.GetYaxis().ChangeLabel(1, -1, 0)
data.SetMarkerStyle(20)
data.SetMarkerSize(1.2)
data.SetLineWidth(2)
data.SetLineColor(ROOT.kBlack)
data.Draw("E SAME")
legend = ROOT.TLegend(0.60, 0.65, 0.92, 0.92)
legend.SetTextFont(42)
legend.SetFillStyle(0)
legend.SetBorderSize(0)
legend.SetTextSize(0.035)
legend.SetTextAlign(32)
legend.AddEntry(data, "Data" ,"lep")
legend.AddEntry(singletop, "Single top + jet", "f")
legend.AddEntry(twtb, "t#bar{t},Wt,t#bar{b}", "f")
legend.AddEntry(wjets, "W+jets", "f")
legend.Draw("SAME")
text = ROOT.TLatex()
text.SetNDC()
text.SetTextFont(72)
text.SetTextSize(0.045)
text.DrawLatex(0.21, 0.86, "ATLAS")
text.SetTextFont(42)
text.DrawLatex(0.21 + 0.16, 0.86, "Open Data")
text.SetTextSize(0.04)
text.DrawLatex(0.21, 0.80, "#sqrt{{s}} = 13 TeV, {:.1f} fb^{{-1}}".format(lumi * lumi_scale / 1000.0))
c.SaveAs("df107_SingleTopAnalysis.png")
print("Saved figure to df107_SingleTopAnalysis.png")
ROOT's RDataFrame offers a high level interface for analyses of data stored in TTree,...
void RunGraphs(std::vector< RResultHandle > handles)
Trigger the event loop of multiple RDataFrames concurrently.
void EnableImplicitMT(UInt_t numthreads=0)
Enable ROOT's implicit multi-threading for all objects and methods that provide an internal paralleli...
A struct which stores the parameters of a TH1D.

 A single top analysis using the ATLAS Open Data release of 2020, with RDataFrame.
A single top analysis using the ATLAS Open Data release of 2020, with RDataFrame.