This macro will perform a scan of the p-values for computing the interval or limit
␛[1mRooFit v3.60 -- Developed by Wouter Verkerke and David Kirkby␛[0m
Copyright (C) 2000-2013 NIKHEF, University of California & Stanford University
All rights reserved, please read http://roofit.sourceforge.net/license.txt
0x5595e6e83780 results/example_combined_GaussExample_model.root
Running HypoTestInverter on the workspace combined
RooWorkspace(combined) combined contents
variables
---------
(Lumi,SigXsecOverSM,alpha_syst1,alpha_syst2,alpha_syst3,binWidth_obs_x_channel1_0,binWidth_obs_x_channel1_1,binWidth_obs_x_channel1_2,channelCat,gamma_stat_channel1_bin_0,gamma_stat_channel1_bin_1,nom_alpha_syst1,nom_alpha_syst2,nom_alpha_syst3,nom_gamma_stat_channel1_bin_0,nom_gamma_stat_channel1_bin_1,nominalLumi,obs_x_channel1,weightVar)
p.d.f.s
-------
RooGaussian::alpha_syst1Constraint[ x=alpha_syst1 mean=nom_alpha_syst1 sigma=1 ] = 1
RooGaussian::alpha_syst2Constraint[ x=alpha_syst2 mean=nom_alpha_syst2 sigma=1 ] = 1
RooGaussian::alpha_syst3Constraint[ x=alpha_syst3 mean=nom_alpha_syst3 sigma=1 ] = 1
RooRealSumPdf::channel1_model[ binWidth_obs_x_channel1_0 * L_x_signal_channel1_overallSyst_x_Exp + binWidth_obs_x_channel1_1 * L_x_background1_channel1_overallSyst_x_StatUncert + binWidth_obs_x_channel1_2 * L_x_background2_channel1_overallSyst_x_StatUncert ] = 220
RooPoisson::gamma_stat_channel1_bin_0_constraint[ x=nom_gamma_stat_channel1_bin_0 mean=gamma_stat_channel1_bin_0_poisMean ] = 0.019943
RooPoisson::gamma_stat_channel1_bin_1_constraint[ x=nom_gamma_stat_channel1_bin_1 mean=gamma_stat_channel1_bin_1_poisMean ] = 0.039861
RooGaussian::lumiConstraint[ x=Lumi mean=nominalLumi sigma=0.1 ] = 1
RooProdPdf::model_channel1[ lumiConstraint * alpha_syst1Constraint * alpha_syst2Constraint * alpha_syst3Constraint * gamma_stat_channel1_bin_0_constraint * gamma_stat_channel1_bin_1_constraint * channel1_model(obs_x_channel1) ] = 0.174888
RooSimultaneous::simPdf[ indexCat=channelCat channel1=model_channel1 ] = 0.174888
functions
--------
RooProduct::L_x_background1_channel1_overallSyst_x_StatUncert[ Lumi * background1_channel1_overallSyst_x_StatUncert ] = 0
RooProduct::L_x_background2_channel1_overallSyst_x_StatUncert[ Lumi * background2_channel1_overallSyst_x_StatUncert ] = 100
RooProduct::L_x_signal_channel1_overallSyst_x_Exp[ Lumi * signal_channel1_overallSyst_x_Exp ] = 10
RooStats::HistFactory::FlexibleInterpVar::background1_channel1_epsilon[ paramList=(alpha_syst2) ] = 1
RooHistFunc::background1_channel1_nominal[ depList=(obs_x_channel1) depList=(obs_x_channel1) ] = 0
RooProduct::background1_channel1_overallSyst_x_Exp[ background1_channel1_nominal * background1_channel1_epsilon ] = 0
RooProduct::background1_channel1_overallSyst_x_StatUncert[ mc_stat_channel1 * background1_channel1_overallSyst_x_Exp ] = 0
RooStats::HistFactory::FlexibleInterpVar::background2_channel1_epsilon[ paramList=(alpha_syst3) ] = 1
RooHistFunc::background2_channel1_nominal[ depList=(obs_x_channel1) depList=(obs_x_channel1) ] = 100
RooProduct::background2_channel1_overallSyst_x_Exp[ background2_channel1_nominal * background2_channel1_epsilon ] = 100
RooProduct::background2_channel1_overallSyst_x_StatUncert[ mc_stat_channel1 * background2_channel1_overallSyst_x_Exp ] = 100
RooProduct::gamma_stat_channel1_bin_0_poisMean[ gamma_stat_channel1_bin_0 * gamma_stat_channel1_bin_0_tau ] = 400
RooProduct::gamma_stat_channel1_bin_1_poisMean[ gamma_stat_channel1_bin_1 * gamma_stat_channel1_bin_1_tau ] = 100
ParamHistFunc::mc_stat_channel1[ ] = 1
RooStats::HistFactory::FlexibleInterpVar::signal_channel1_epsilon[ paramList=(alpha_syst1) ] = 1
RooHistFunc::signal_channel1_nominal[ depList=(obs_x_channel1) depList=(obs_x_channel1) ] = 10
RooProduct::signal_channel1_overallNorm_x_sigma_epsilon[ SigXsecOverSM * signal_channel1_epsilon ] = 1
RooProduct::signal_channel1_overallSyst_x_Exp[ signal_channel1_nominal * signal_channel1_overallNorm_x_sigma_epsilon ] = 10
datasets
--------
RooDataSet::asimovData(obs_x_channel1,weightVar,channelCat)
RooDataSet::obsData(channelCat,obs_x_channel1)
embedded datasets (in pdfs and functions)
-----------------------------------------
RooDataHist::signal_channel1nominalDHist(obs_x_channel1)
RooDataHist::background1_channel1nominalDHist(obs_x_channel1)
RooDataHist::background2_channel1nominalDHist(obs_x_channel1)
parameter snapshots
-------------------
NominalParamValues = (nom_alpha_syst2=0[C],nom_alpha_syst3=0[C],nom_gamma_stat_channel1_bin_0=400[C],nom_gamma_stat_channel1_bin_1=100[C],weightVar=0,obs_x_channel1=1.75,Lumi=1[C],nominalLumi=1[C],alpha_syst1=0[C],nom_alpha_syst1=0[C],alpha_syst2=0,alpha_syst3=0,gamma_stat_channel1_bin_0=1,gamma_stat_channel1_bin_1=1,SigXsecOverSM=1,binWidth_obs_x_channel1_0=2[C],binWidth_obs_x_channel1_1=2[C],binWidth_obs_x_channel1_2=2[C])
named sets
----------
ModelConfig_GlobalObservables:(nom_alpha_syst2,nom_alpha_syst3,nom_gamma_stat_channel1_bin_0,nom_gamma_stat_channel1_bin_1)
ModelConfig_NuisParams:(alpha_syst2,alpha_syst3,gamma_stat_channel1_bin_0,gamma_stat_channel1_bin_1)
ModelConfig_Observables:(obs_x_channel1,weightVar,channelCat)
ModelConfig_POI:(SigXsecOverSM)
globalObservables:(nom_alpha_syst2,nom_alpha_syst3,nom_gamma_stat_channel1_bin_0,nom_gamma_stat_channel1_bin_1)
observables:(obs_x_channel1,weightVar,channelCat)
generic objects
---------------
RooStats::ModelConfig::ModelConfig
Using data set obsData
StandardHypoTestInvDemo : POI initial value: SigXsecOverSM = 1
[#1] INFO:InputArguments -- HypoTestInverter ---- Input models:
using as S+B (null) model : ModelConfig
using as B (alternate) model : ModelConfig_with_poi_0
Doing a fixed scan in interval : 0 , 5
[#1] INFO:Eval -- HypoTestInverter::GetInterval - run a fixed scan
[#0] WARNING:InputArguments -- HypoTestInverter::RunFixedScan - xMax > upper bound, use xmax = 3
[#1] INFO:ObjectHandling -- RooWorkspace::saveSnaphot(combined) replacing previous snapshot with name ModelConfig__snapshot
[#0] PROGRESS:Eval -- Running for SigXsecOverSM = 0
=== Using the following for ModelConfig ===
Observables: RooArgSet:: = (obs_x_channel1,weightVar,channelCat)
Parameters of Interest: RooArgSet:: = (SigXsecOverSM)
Nuisance Parameters: RooArgSet:: = (alpha_syst2,alpha_syst3,gamma_stat_channel1_bin_0,gamma_stat_channel1_bin_1)
Global Observables: RooArgSet:: = (nom_alpha_syst2,nom_alpha_syst3,nom_gamma_stat_channel1_bin_0,nom_gamma_stat_channel1_bin_1)
PDF: RooSimultaneous::simPdf[ indexCat=channelCat channel1=model_channel1 ] = 0.158989
Snapshot:
1) 0x5595e7a5c980 RooRealVar:: SigXsecOverSM = 0 L(0 - 3) "SigXsecOverSM"
=== Using the following for ModelConfig_with_poi_0 ===
Observables: RooArgSet:: = (obs_x_channel1,weightVar,channelCat)
Parameters of Interest: RooArgSet:: = (SigXsecOverSM)
Nuisance Parameters: RooArgSet:: = (alpha_syst2,alpha_syst3,gamma_stat_channel1_bin_0,gamma_stat_channel1_bin_1)
Global Observables: RooArgSet:: = (nom_alpha_syst2,nom_alpha_syst3,nom_gamma_stat_channel1_bin_0,nom_gamma_stat_channel1_bin_1)
PDF: RooSimultaneous::simPdf[ indexCat=channelCat channel1=model_channel1 ] = 0.158989
Snapshot:
1) 0x5595e7a5c980 RooRealVar:: SigXsecOverSM = 0 L(0 - 3) "SigXsecOverSM"
[#0] PROGRESS:Generation -- Test Statistic on data: 0
[#1] INFO:InputArguments -- Profiling conditional MLEs for Null.
[#1] INFO:InputArguments -- Using a ToyMCSampler. Now configuring for Null.
[#0] PROGRESS:Generation -- generated toys: 500 / 1000
[#1] INFO:InputArguments -- Profiling conditional MLEs for Alt.
[#1] INFO:InputArguments -- Using a ToyMCSampler. Now configuring for Alt.
[#0] PROGRESS:Eval -- P values for SigXsecOverSM = 0
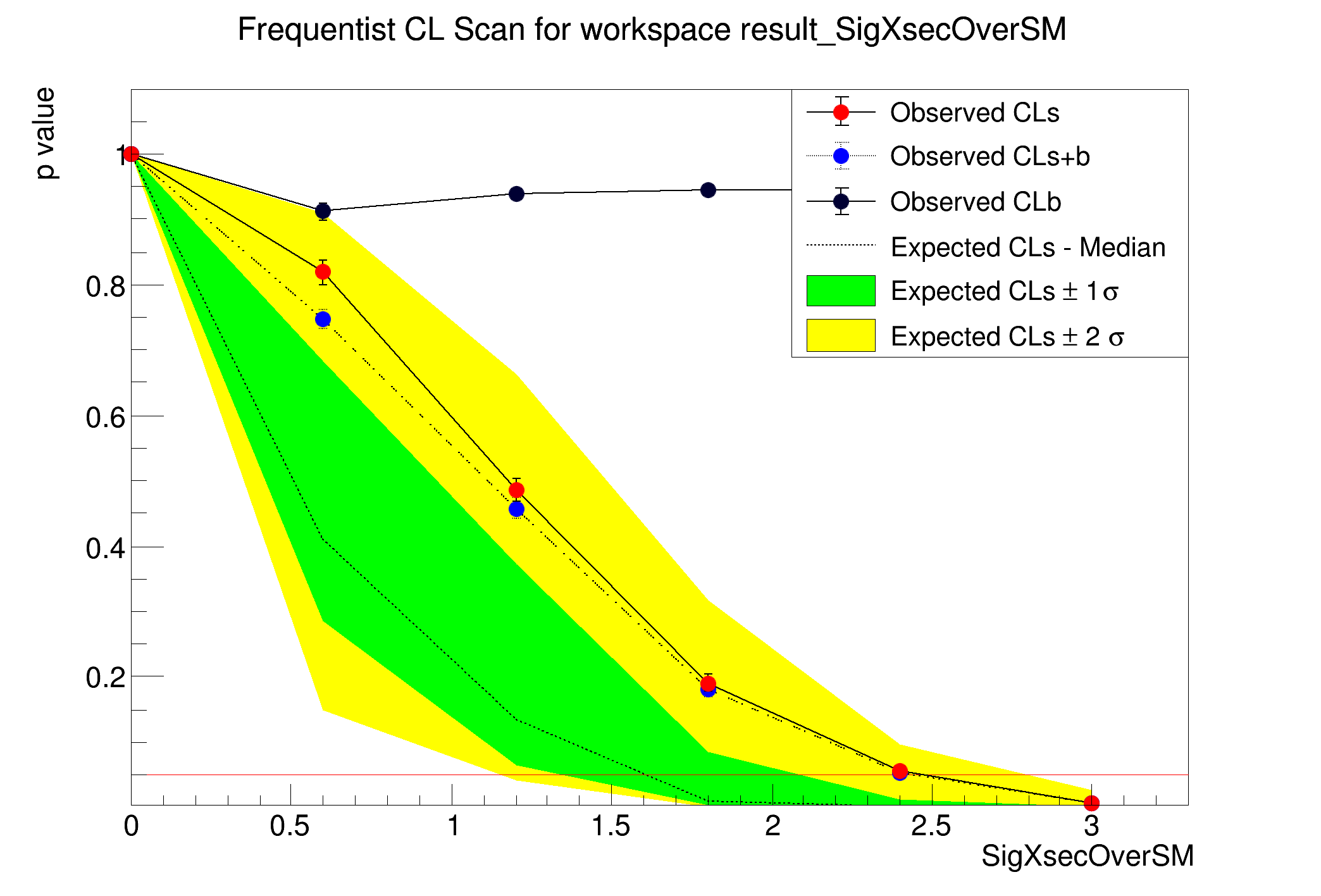
CLs = 1 +/- 0
CLb = 1 +/- 0
CLsplusb = 1 +/- 0
[#1] INFO:ObjectHandling -- RooWorkspace::saveSnaphot(combined) replacing previous snapshot with name ModelConfig__snapshot
[#0] PROGRESS:Eval -- Running for SigXsecOverSM = 0.6
=== Using the following for ModelConfig ===
Observables: RooArgSet:: = (obs_x_channel1,weightVar,channelCat)
Parameters of Interest: RooArgSet:: = (SigXsecOverSM)
Nuisance Parameters: RooArgSet:: = (alpha_syst2,alpha_syst3,gamma_stat_channel1_bin_0,gamma_stat_channel1_bin_1)
Global Observables: RooArgSet:: = (nom_alpha_syst2,nom_alpha_syst3,nom_gamma_stat_channel1_bin_0,nom_gamma_stat_channel1_bin_1)
PDF: RooSimultaneous::simPdf[ indexCat=channelCat channel1=model_channel1 ] = 0.168529
Snapshot:
1) 0x5595e7cfd5c0 RooRealVar:: SigXsecOverSM = 0.6 L(0 - 3) "SigXsecOverSM"
=== Using the following for ModelConfig_with_poi_0 ===
Observables: RooArgSet:: = (obs_x_channel1,weightVar,channelCat)
Parameters of Interest: RooArgSet:: = (SigXsecOverSM)
Nuisance Parameters: RooArgSet:: = (alpha_syst2,alpha_syst3,gamma_stat_channel1_bin_0,gamma_stat_channel1_bin_1)
Global Observables: RooArgSet:: = (nom_alpha_syst2,nom_alpha_syst3,nom_gamma_stat_channel1_bin_0,nom_gamma_stat_channel1_bin_1)
PDF: RooSimultaneous::simPdf[ indexCat=channelCat channel1=model_channel1 ] = 0.168529
Snapshot:
1) 0x5595e7cfd5c0 RooRealVar:: SigXsecOverSM = 0 L(0 - 3) "SigXsecOverSM"
[#0] PROGRESS:Generation -- Test Statistic on data: -2.35222
[#1] INFO:InputArguments -- Profiling conditional MLEs for Null.
[#1] INFO:InputArguments -- Using a ToyMCSampler. Now configuring for Null.
[#0] PROGRESS:Generation -- generated toys: 500 / 1000
[#1] INFO:InputArguments -- Profiling conditional MLEs for Alt.
[#1] INFO:InputArguments -- Using a ToyMCSampler. Now configuring for Alt.
[#0] PROGRESS:Eval -- P values for SigXsecOverSM = 0.6
CLs = 0.819079 +/- 0.0188863
CLb = 0.912 +/- 0.0126693
CLsplusb = 0.747 +/- 0.0137474
[#1] INFO:ObjectHandling -- RooWorkspace::saveSnaphot(combined) replacing previous snapshot with name ModelConfig__snapshot
[#0] PROGRESS:Eval -- Running for SigXsecOverSM = 1.2
=== Using the following for ModelConfig ===
Observables: RooArgSet:: = (obs_x_channel1,weightVar,channelCat)
Parameters of Interest: RooArgSet:: = (SigXsecOverSM)
Nuisance Parameters: RooArgSet:: = (alpha_syst2,alpha_syst3,gamma_stat_channel1_bin_0,gamma_stat_channel1_bin_1)
Global Observables: RooArgSet:: = (nom_alpha_syst2,nom_alpha_syst3,nom_gamma_stat_channel1_bin_0,nom_gamma_stat_channel1_bin_1)
PDF: RooSimultaneous::simPdf[ indexCat=channelCat channel1=model_channel1 ] = 0.178068
Snapshot:
1) 0x5595e7d52b50 RooRealVar:: SigXsecOverSM = 1.2 L(0 - 3) "SigXsecOverSM"
=== Using the following for ModelConfig_with_poi_0 ===
Observables: RooArgSet:: = (obs_x_channel1,weightVar,channelCat)
Parameters of Interest: RooArgSet:: = (SigXsecOverSM)
Nuisance Parameters: RooArgSet:: = (alpha_syst2,alpha_syst3,gamma_stat_channel1_bin_0,gamma_stat_channel1_bin_1)
Global Observables: RooArgSet:: = (nom_alpha_syst2,nom_alpha_syst3,nom_gamma_stat_channel1_bin_0,nom_gamma_stat_channel1_bin_1)
PDF: RooSimultaneous::simPdf[ indexCat=channelCat channel1=model_channel1 ] = 0.178068
Snapshot:
1) 0x5595e7d52b50 RooRealVar:: SigXsecOverSM = 0 L(0 - 3) "SigXsecOverSM"
[#0] PROGRESS:Generation -- Test Statistic on data: -2.9364
[#1] INFO:InputArguments -- Profiling conditional MLEs for Null.
[#1] INFO:InputArguments -- Using a ToyMCSampler. Now configuring for Null.
[#0] PROGRESS:Generation -- generated toys: 500 / 1000
[#1] INFO:InputArguments -- Profiling conditional MLEs for Alt.
[#1] INFO:InputArguments -- Using a ToyMCSampler. Now configuring for Alt.
[#0] PROGRESS:Eval -- P values for SigXsecOverSM = 1.2
CLs = 0.48617 +/- 0.0176356
CLb = 0.94 +/- 0.0106207
CLsplusb = 0.457 +/- 0.0157528
[#1] INFO:ObjectHandling -- RooWorkspace::saveSnaphot(combined) replacing previous snapshot with name ModelConfig__snapshot
[#0] PROGRESS:Eval -- Running for SigXsecOverSM = 1.8
=== Using the following for ModelConfig ===
Observables: RooArgSet:: = (obs_x_channel1,weightVar,channelCat)
Parameters of Interest: RooArgSet:: = (SigXsecOverSM)
Nuisance Parameters: RooArgSet:: = (alpha_syst2,alpha_syst3,gamma_stat_channel1_bin_0,gamma_stat_channel1_bin_1)
Global Observables: RooArgSet:: = (nom_alpha_syst2,nom_alpha_syst3,nom_gamma_stat_channel1_bin_0,nom_gamma_stat_channel1_bin_1)
PDF: RooSimultaneous::simPdf[ indexCat=channelCat channel1=model_channel1 ] = 0.187607
Snapshot:
1) 0x5595e7c53a80 RooRealVar:: SigXsecOverSM = 1.8 L(0 - 3) "SigXsecOverSM"
=== Using the following for ModelConfig_with_poi_0 ===
Observables: RooArgSet:: = (obs_x_channel1,weightVar,channelCat)
Parameters of Interest: RooArgSet:: = (SigXsecOverSM)
Nuisance Parameters: RooArgSet:: = (alpha_syst2,alpha_syst3,gamma_stat_channel1_bin_0,gamma_stat_channel1_bin_1)
Global Observables: RooArgSet:: = (nom_alpha_syst2,nom_alpha_syst3,nom_gamma_stat_channel1_bin_0,nom_gamma_stat_channel1_bin_1)
PDF: RooSimultaneous::simPdf[ indexCat=channelCat channel1=model_channel1 ] = 0.187607
Snapshot:
1) 0x5595e7c53a80 RooRealVar:: SigXsecOverSM = 0 L(0 - 3) "SigXsecOverSM"
[#0] PROGRESS:Generation -- Test Statistic on data: -2.05075
[#1] INFO:InputArguments -- Profiling conditional MLEs for Null.
[#1] INFO:InputArguments -- Using a ToyMCSampler. Now configuring for Null.
[#0] PROGRESS:Generation -- generated toys: 500 / 1000
[#1] INFO:InputArguments -- Profiling conditional MLEs for Alt.
[#1] INFO:InputArguments -- Using a ToyMCSampler. Now configuring for Alt.
[#0] PROGRESS:Eval -- P values for SigXsecOverSM = 1.8
CLs = 0.190678 +/- 0.0130363
CLb = 0.944 +/- 0.0102824
CLsplusb = 0.18 +/- 0.0121491
[#1] INFO:ObjectHandling -- RooWorkspace::saveSnaphot(combined) replacing previous snapshot with name ModelConfig__snapshot
[#0] PROGRESS:Eval -- Running for SigXsecOverSM = 2.4
=== Using the following for ModelConfig ===
Observables: RooArgSet:: = (obs_x_channel1,weightVar,channelCat)
Parameters of Interest: RooArgSet:: = (SigXsecOverSM)
Nuisance Parameters: RooArgSet:: = (alpha_syst2,alpha_syst3,gamma_stat_channel1_bin_0,gamma_stat_channel1_bin_1)
Global Observables: RooArgSet:: = (nom_alpha_syst2,nom_alpha_syst3,nom_gamma_stat_channel1_bin_0,nom_gamma_stat_channel1_bin_1)
PDF: RooSimultaneous::simPdf[ indexCat=channelCat channel1=model_channel1 ] = 0.197147
Snapshot:
1) 0x5595e7cfd5c0 RooRealVar:: SigXsecOverSM = 2.4 L(0 - 3) "SigXsecOverSM"
=== Using the following for ModelConfig_with_poi_0 ===
Observables: RooArgSet:: = (obs_x_channel1,weightVar,channelCat)
Parameters of Interest: RooArgSet:: = (SigXsecOverSM)
Nuisance Parameters: RooArgSet:: = (alpha_syst2,alpha_syst3,gamma_stat_channel1_bin_0,gamma_stat_channel1_bin_1)
Global Observables: RooArgSet:: = (nom_alpha_syst2,nom_alpha_syst3,nom_gamma_stat_channel1_bin_0,nom_gamma_stat_channel1_bin_1)
PDF: RooSimultaneous::simPdf[ indexCat=channelCat channel1=model_channel1 ] = 0.197147
Snapshot:
1) 0x5595e7cfd5c0 RooRealVar:: SigXsecOverSM = 0 L(0 - 3) "SigXsecOverSM"
[#0] PROGRESS:Generation -- Test Statistic on data: 0.0783908
[#1] INFO:InputArguments -- Profiling conditional MLEs for Null.
[#1] INFO:InputArguments -- Using a ToyMCSampler. Now configuring for Null.
[#0] PROGRESS:Generation -- generated toys: 500 / 1000
[#1] INFO:InputArguments -- Profiling conditional MLEs for Alt.
[#1] INFO:InputArguments -- Using a ToyMCSampler. Now configuring for Alt.
[#0] PROGRESS:Eval -- P values for SigXsecOverSM = 2.4
CLs = 0.0550847 +/- 0.00746178
CLb = 0.944 +/- 0.0102824
CLsplusb = 0.052 +/- 0.00702111
[#1] INFO:ObjectHandling -- RooWorkspace::saveSnaphot(combined) replacing previous snapshot with name ModelConfig__snapshot
[#0] PROGRESS:Eval -- Running for SigXsecOverSM = 3
=== Using the following for ModelConfig ===
Observables: RooArgSet:: = (obs_x_channel1,weightVar,channelCat)
Parameters of Interest: RooArgSet:: = (SigXsecOverSM)
Nuisance Parameters: RooArgSet:: = (alpha_syst2,alpha_syst3,gamma_stat_channel1_bin_0,gamma_stat_channel1_bin_1)
Global Observables: RooArgSet:: = (nom_alpha_syst2,nom_alpha_syst3,nom_gamma_stat_channel1_bin_0,nom_gamma_stat_channel1_bin_1)
PDF: RooSimultaneous::simPdf[ indexCat=channelCat channel1=model_channel1 ] = 0.206686
Snapshot:
1) 0x5595e7d52b50 RooRealVar:: SigXsecOverSM = 3 L(0 - 3) "SigXsecOverSM"
=== Using the following for ModelConfig_with_poi_0 ===
Observables: RooArgSet:: = (obs_x_channel1,weightVar,channelCat)
Parameters of Interest: RooArgSet:: = (SigXsecOverSM)
Nuisance Parameters: RooArgSet:: = (alpha_syst2,alpha_syst3,gamma_stat_channel1_bin_0,gamma_stat_channel1_bin_1)
Global Observables: RooArgSet:: = (nom_alpha_syst2,nom_alpha_syst3,nom_gamma_stat_channel1_bin_0,nom_gamma_stat_channel1_bin_1)
PDF: RooSimultaneous::simPdf[ indexCat=channelCat channel1=model_channel1 ] = 0.206686
Snapshot:
1) 0x5595e7d52b50 RooRealVar:: SigXsecOverSM = 0 L(0 - 3) "SigXsecOverSM"
[#0] PROGRESS:Generation -- Test Statistic on data: 3.27476
[#1] INFO:InputArguments -- Profiling conditional MLEs for Null.
[#1] INFO:InputArguments -- Using a ToyMCSampler. Now configuring for Null.
[#0] PROGRESS:Generation -- generated toys: 500 / 1000
[#1] INFO:InputArguments -- Profiling conditional MLEs for Alt.
[#1] INFO:InputArguments -- Using a ToyMCSampler. Now configuring for Alt.
[#0] PROGRESS:Eval -- P values for SigXsecOverSM = 3
CLs = 0.00535332 +/- 0.00238893
CLb = 0.934 +/- 0.0111035
CLsplusb = 0.005 +/- 0.00223047
Time to perform limit scan
Real time 0:00:13, CP time 13.470
The computed upper limit is: 2.46135 +/- 0.0596845
Expected upper limits, using the B (alternate) model :
expected limit (median) 1.60988
expected limit (-1 sig) 1.34011
expected limit (+1 sig) 2.09019
expected limit (-2 sig) 1.14968
expected limit (+2 sig) 2.79445
[#0] WARNING:Plotting -- Could not determine xmin and xmax of sampling distribution that was given to plot.
[#0] WARNING:Plotting -- Could not determine xmin and xmax of sampling distribution that was given to plot.
#include <cassert>
struct HypoTestInvOptions {
bool plotHypoTestResult = true;
bool writeResult = true;
bool optimize = true;
bool useVectorStore = true;
bool generateBinned = false;
bool noSystematics = false;
double nToysRatio = 2;
double maxPOI = -1;
bool useProof = false;
int nworkers = 0;
bool enableDetailedOutput = false;
bool rebuild = false;
int nToyToRebuild = 100;
int rebuildParamValues=0;
int initialFit = -1;
int randomSeed = -1;
int nAsimovBins = 0;
bool reuseAltToys = false;
double confLevel = 0.95;
std::string minimizerType = "";
std::string massValue = "";
int printLevel = 0;
bool useNLLOffset = false;
};
HypoTestInvOptions optHTInv;
class HypoTestInvTool{
public:
HypoTestInvTool();
~HypoTestInvTool(){};
HypoTestInverterResult *
const char * modelSBName, const char * modelBName,
const char * dataName,
int type,
int testStatType,
bool useCLs,
int npoints, double poimin, double poimax, int ntoys,
bool useNumberCounting = false,
const char * nuisPriorName = 0);
void
AnalyzeResult( HypoTestInverterResult *
r,
int calculatorType,
int testStatType,
bool useCLs,
int npoints,
const char * fileNameBase = 0 );
void SetParameter(
const char *
name,
const char * value);
void SetParameter(
const char *
name,
bool value);
void SetParameter(
const char *
name,
int value);
void SetParameter(
const char *
name,
double value);
private:
bool mPlotHypoTestResult;
bool mWriteResult;
bool mOptimize;
bool mUseVectorStore;
bool mGenerateBinned;
bool mUseProof;
bool mRebuild;
bool mReuseAltToys;
bool mEnableDetOutput;
int mNWorkers;
int mNToyToRebuild;
int mRebuildParamValues;
int mPrintLevel;
int mInitialFit;
int mRandomSeed;
double mNToysRatio;
double mMaxPoi;
int mAsimovBins;
std::string mMassValue;
std::string mMinimizerType;
};
}
RooStats::HypoTestInvTool::HypoTestInvTool() : mPlotHypoTestResult(true),
mWriteResult(false),
mOptimize(true),
mUseVectorStore(true),
mGenerateBinned(false),
mUseProof(false),
mEnableDetOutput(false),
mRebuild(false),
mReuseAltToys(false),
mNWorkers(4),
mNToyToRebuild(100),
mRebuildParamValues(0),
mPrintLevel(0),
mInitialFit(-1),
mRandomSeed(-1),
mNToysRatio(2),
mMaxPoi(-1),
mAsimovBins(0),
mMassValue(""),
mMinimizerType(""),
mResultFileName() {
}
void
RooStats::HypoTestInvTool::SetParameter(
const char *
name,
bool value){
std::string s_name(
name);
if (s_name.find("PlotHypoTestResult") != std::string::npos) mPlotHypoTestResult = value;
if (s_name.find("WriteResult") != std::string::npos) mWriteResult = value;
if (s_name.find("Optimize") != std::string::npos) mOptimize = value;
if (s_name.find("UseVectorStore") != std::string::npos) mUseVectorStore = value;
if (s_name.find("GenerateBinned") != std::string::npos) mGenerateBinned = value;
if (s_name.find("UseProof") != std::string::npos) mUseProof = value;
if (s_name.find("EnableDetailedOutput") != std::string::npos) mEnableDetOutput = value;
if (s_name.find("Rebuild") != std::string::npos) mRebuild = value;
if (s_name.find("ReuseAltToys") != std::string::npos) mReuseAltToys = value;
return;
}
void
RooStats::HypoTestInvTool::SetParameter(
const char *
name,
int value){
std::string s_name(
name);
if (s_name.find("NWorkers") != std::string::npos) mNWorkers = value;
if (s_name.find("NToyToRebuild") != std::string::npos) mNToyToRebuild = value;
if (s_name.find("RebuildParamValues") != std::string::npos) mRebuildParamValues = value;
if (s_name.find("PrintLevel") != std::string::npos) mPrintLevel = value;
if (s_name.find("InitialFit") != std::string::npos) mInitialFit = value;
if (s_name.find("RandomSeed") != std::string::npos) mRandomSeed = value;
if (s_name.find("AsimovBins") != std::string::npos) mAsimovBins = value;
return;
}
void
RooStats::HypoTestInvTool::SetParameter(
const char *
name,
double value){
std::string s_name(
name);
if (s_name.find("NToysRatio") != std::string::npos) mNToysRatio = value;
if (s_name.find("MaxPOI") != std::string::npos) mMaxPoi = value;
return;
}
void
RooStats::HypoTestInvTool::SetParameter(
const char *
name,
const char * value){
std::string s_name(
name);
if (s_name.find("MassValue") != std::string::npos) mMassValue.assign(value);
if (s_name.find("MinimizerType") != std::string::npos) mMinimizerType.assign(value);
if (s_name.find("ResultFileName") != std::string::npos) mResultFileName = value;
return;
}
void
StandardHypoTestInvDemo(const char * infile = 0,
const char * wsName = "combined",
const char * modelSBName = "ModelConfig",
const char * modelBName = "",
const char * dataName = "obsData",
int calculatorType = 0,
int testStatType = 0,
bool useCLs = true ,
int npoints = 6,
double poimin = 0,
double poimax = 5,
int ntoys=1000,
bool useNumberCounting = false,
const char * nuisPriorName = 0){
if (filename.IsNull()) {
filename = "results/example_combined_GaussExample_model.root";
if (!fileExist) {
#ifdef _WIN32
cout << "HistFactory file cannot be generated on Windows - exit" << endl;
return;
#endif
cout <<"will run standard hist2workspace example"<<endl;
gROOT->ProcessLine(
".! prepareHistFactory .");
gROOT->ProcessLine(
".! hist2workspace config/example.xml");
cout <<"\n\n---------------------"<<endl;
cout <<"Done creating example input"<<endl;
cout <<"---------------------\n\n"<<endl;
}
}
else
filename = infile;
cout <<"StandardRooStatsDemoMacro: Input file " << filename << " is not found" << endl;
return;
}
HypoTestInvTool calc;
calc.SetParameter("PlotHypoTestResult", optHTInv.plotHypoTestResult);
calc.SetParameter("WriteResult", optHTInv.writeResult);
calc.SetParameter("Optimize", optHTInv.optimize);
calc.SetParameter("UseVectorStore", optHTInv.useVectorStore);
calc.SetParameter("GenerateBinned", optHTInv.generateBinned);
calc.SetParameter("NToysRatio", optHTInv.nToysRatio);
calc.SetParameter("MaxPOI", optHTInv.maxPOI);
calc.SetParameter("UseProof", optHTInv.useProof);
calc.SetParameter("EnableDetailedOutput", optHTInv.enableDetailedOutput);
calc.SetParameter("NWorkers", optHTInv.nworkers);
calc.SetParameter("Rebuild", optHTInv.rebuild);
calc.SetParameter("ReuseAltToys", optHTInv.reuseAltToys);
calc.SetParameter("NToyToRebuild", optHTInv.nToyToRebuild);
calc.SetParameter("RebuildParamValues", optHTInv.rebuildParamValues);
calc.SetParameter("MassValue", optHTInv.massValue.c_str());
calc.SetParameter("MinimizerType", optHTInv.minimizerType.c_str());
calc.SetParameter("PrintLevel", optHTInv.printLevel);
calc.SetParameter("InitialFit", optHTInv.initialFit);
calc.SetParameter("ResultFileName", optHTInv.resultFileName);
calc.SetParameter("RandomSeed", optHTInv.randomSeed);
calc.SetParameter("AsimovBins", optHTInv.nAsimovBins);
HypoTestInverterResult *
r = 0;
std::cout << w << "\t" << filename << std::endl;
if (w != NULL) {
r = calc.RunInverter(w, modelSBName, modelBName,
dataName, calculatorType, testStatType, useCLs,
npoints, poimin, poimax,
ntoys, useNumberCounting, nuisPriorName );
std::cerr << "Error running the HypoTestInverter - Exit " << std::endl;
return;
}
}
else {
std::cout << "Reading an HypoTestInverterResult with name " << wsName << " from file " << filename << std::endl;
r =
dynamic_cast<HypoTestInverterResult*
>(
file->Get(wsName) );
std::cerr << "File " << filename << " does not contain a workspace or an HypoTestInverterResult - Exit "
<< std::endl;
return;
}
}
calc.AnalyzeResult(
r, calculatorType, testStatType, useCLs, npoints, infile );
return;
}
void
RooStats::HypoTestInvTool::AnalyzeResult( HypoTestInverterResult *
r,
int calculatorType,
int testStatType,
bool useCLs,
int npoints,
const char * fileNameBase ){
double lowerLimit = 0;
double llError = 0;
#if defined ROOT_SVN_VERSION && ROOT_SVN_VERSION >= 44126
lowerLimit =
r->LowerLimit();
llError =
r->LowerLimitEstimatedError();
}
#else
lowerLimit =
r->LowerLimit();
llError =
r->LowerLimitEstimatedError();
#endif
double upperLimit =
r->UpperLimit();
double ulError =
r->UpperLimitEstimatedError();
if (lowerLimit < upperLimit*(1.- 1.E-4) && lowerLimit != 0)
std::cout << "The computed lower limit is: " << lowerLimit << " +/- " << llError << std::endl;
std::cout << "The computed upper limit is: " << upperLimit << " +/- " << ulError << std::endl;
std::cout << "Expected upper limits, using the B (alternate) model : " << std::endl;
std::cout <<
" expected limit (median) " <<
r->GetExpectedUpperLimit(0) << std::endl;
std::cout <<
" expected limit (-1 sig) " <<
r->GetExpectedUpperLimit(-1) << std::endl;
std::cout <<
" expected limit (+1 sig) " <<
r->GetExpectedUpperLimit(1) << std::endl;
std::cout <<
" expected limit (-2 sig) " <<
r->GetExpectedUpperLimit(-2) << std::endl;
std::cout <<
" expected limit (+2 sig) " <<
r->GetExpectedUpperLimit(2) << std::endl;
if (mEnableDetOutput) {
mWriteResult=true;
Info(
"StandardHypoTestInvDemo",
"detailed output will be written in output result file");
}
if (
r != NULL && mWriteResult) {
const char * calcType = (calculatorType == 0) ? "Freq" : (calculatorType == 1) ? "Hybr" : "Asym";
const char * limitType = (useCLs) ? "CLs" : "Cls+b";
const char * scanType = (npoints < 0) ? "auto" : "grid";
if (mResultFileName.IsNull()) {
mResultFileName =
TString::Format(
"%s_%s_%s_ts%d_",calcType,limitType,scanType,testStatType);
if (mMassValue.size()>0) {
mResultFileName += mMassValue.c_str();
mResultFileName += "_";
}
}
TString uldistFile =
"RULDist.root";
if (existULDist) {
if (fileULDist) ulDist= fileULDist->
Get(
"RULDist");
}
TFile * fileOut =
new TFile(mResultFileName,
"RECREATE");
if (ulDist) ulDist->
Write();
Info(
"StandardHypoTestInvDemo",
"HypoTestInverterResult has been written in the file %s",mResultFileName.Data());
}
std::string typeName = "";
if (calculatorType == 0 )
typeName = "Frequentist";
if (calculatorType == 1 )
typeName = "Hybrid";
else if (calculatorType == 2 || calculatorType == 3) {
typeName = "Asymptotic";
mPlotHypoTestResult = false;
}
HypoTestInverterPlot *plot =
new HypoTestInverterPlot(
"HTI_Result_Plot",plotTitle,
r);
plot->Draw("CLb 2CL");
const int nEntries =
r->ArraySize();
if (mPlotHypoTestResult) {
if (nEntries > 1) {
}
for (int i=0; i<nEntries; i++) {
if (nEntries > 1)
c2->cd(i+1);
SamplingDistPlot * pl = plot->MakeTestStatPlot(i);
pl->SetLogYaxis(true);
pl->Draw();
}
}
}
HypoTestInverterResult *
const char * modelSBName, const char * modelBName,
const char * dataName,
int type,
int testStatType,
bool useCLs, int npoints, double poimin, double poimax,
int ntoys,
bool useNumberCounting,
const char * nuisPriorName ){
std::cout <<
"Running HypoTestInverter on the workspace " << w->
GetName() << std::endl;
Error(
"StandardHypoTestDemo",
"Not existing data %s",dataName);
return 0;
}
else
std::cout << "Using data set " << dataName << std::endl;
if (mUseVectorStore) {
data->convertToVectorStore() ;
}
ModelConfig* bModel = (ModelConfig*) w->
obj(modelBName);
ModelConfig* sbModel = (ModelConfig*) w->
obj(modelSBName);
if (!sbModel) {
Error(
"StandardHypoTestDemo",
"Not existing ModelConfig %s",modelSBName);
return 0;
}
if (!sbModel->GetPdf()) {
Error(
"StandardHypoTestDemo",
"Model %s has no pdf ",modelSBName);
return 0;
}
if (!sbModel->GetParametersOfInterest()) {
Error(
"StandardHypoTestDemo",
"Model %s has no poi ",modelSBName);
return 0;
}
if (!sbModel->GetObservables()) {
Error(
"StandardHypoTestInvDemo",
"Model %s has no observables ",modelSBName);
return 0;
}
if (!sbModel->GetSnapshot() ) {
Info(
"StandardHypoTestInvDemo",
"Model %s has no snapshot - make one using model poi",modelSBName);
sbModel->SetSnapshot( *sbModel->GetParametersOfInterest() );
}
if (optHTInv.noSystematics) {
const RooArgSet * nuisPar = sbModel->GetNuisanceParameters();
if (nuisPar && nuisPar->
getSize() > 0) {
std::cout << "StandardHypoTestInvDemo" << " - Switch off all systematics by setting them constant to their initial values" << std::endl;
}
if (bModel) {
const RooArgSet * bnuisPar = bModel->GetNuisanceParameters();
if (bnuisPar)
}
}
if (!bModel || bModel == sbModel) {
Info(
"StandardHypoTestInvDemo",
"The background model %s does not exist",modelBName);
Info(
"StandardHypoTestInvDemo",
"Copy it from ModelConfig %s and set POI to zero",modelSBName);
bModel = (ModelConfig*) sbModel->Clone();
if (!var) return 0;
double oldval = var->
getVal();
}
else {
if (!bModel->GetSnapshot() ) {
Info(
"StandardHypoTestInvDemo",
"Model %s has no snapshot - make one using model poi and 0 values ",modelBName);
if (var) {
double oldval = var->
getVal();
}
else {
Error(
"StandardHypoTestInvDemo",
"Model %s has no valid poi",modelBName);
return 0;
}
}
}
bool hasNuisParam = (sbModel->GetNuisanceParameters() && sbModel->GetNuisanceParameters()->getSize() > 0);
bool hasGlobalObs = (sbModel->GetGlobalObservables() && sbModel->GetGlobalObservables()->getSize() > 0);
if (hasNuisParam && !hasGlobalObs ) {
if (constrPdf) {
Warning(
"StandardHypoTestInvDemo",
"Model %s has nuisance parameters but no global observables associated",sbModel->GetName());
Warning(
"StandardHypoTestInvDemo",
"\tThe effect of the nuisance parameters will not be treated correctly ");
}
}
}
delete allParams;
const RooArgSet * poiSet = sbModel->GetParametersOfInterest();
std::cout <<
"StandardHypoTestInvDemo : POI initial value: " << poi->
GetName() <<
" = " << poi->
getVal() << std::endl;
bool doFit = mInitialFit;
if (testStatType == 0 && mInitialFit == -1) doFit = false;
if (
type == 3 && mInitialFit == -1) doFit =
false;
double poihat = 0;
else
Info(
"StandardHypoTestInvDemo",
"Using %s as minimizer for computing the test statistic",
if (doFit) {
Info(
"StandardHypoTestInvDemo",
" Doing a first fit to the observed data ");
if (sbModel->GetNuisanceParameters() ) constrainParams.
add(*sbModel->GetNuisanceParameters());
Warning(
"StandardHypoTestInvDemo",
"Fit to the model failed - try with strategy 1 and perform first an Hesse computation");
}
Warning(
"StandardHypoTestInvDemo",
" Fit still failed - continue anyway.....");
std::cout <<
"StandardHypoTestInvDemo - Best Fit value : " << poi->
GetName() <<
" = " << poihat <<
" +/- " << poi->
getError() << std::endl;
std::cout <<
"Time for fitting : "; tw.
Print();
sbModel->SetSnapshot(*sbModel->GetParametersOfInterest());
std::cout << "StandardHypoTestInvo: snapshot of S+B Model " << sbModel->GetName()
<< " is set to the best fit value" << std::endl;
}
if (testStatType == 0) {
if (!doFit)
Info(
"StandardHypoTestInvDemo",
"Using LEP test statistic - an initial fit is not done and the TS will use the nuisances at the model value");
else
Info(
"StandardHypoTestInvDemo",
"Using LEP test statistic - an initial fit has been done and the TS will use the nuisances at the best fit value");
}
SimpleLikelihoodRatioTestStat slrts(*sbModel->GetPdf(),*bModel->GetPdf());
RooArgSet nullParams(*sbModel->GetSnapshot());
if (sbModel->GetNuisanceParameters()) nullParams.add(*sbModel->GetNuisanceParameters());
if (sbModel->GetSnapshot()) slrts.SetNullParameters(nullParams);
if (bModel->GetNuisanceParameters()) altParams.add(*bModel->GetNuisanceParameters());
if (bModel->GetSnapshot()) slrts.SetAltParameters(altParams);
if (mEnableDetOutput) slrts.EnableDetailedOutput();
RatioOfProfiledLikelihoodsTestStat
ropl(*sbModel->GetPdf(), *bModel->GetPdf(), bModel->GetSnapshot());
ropl.SetSubtractMLE(false);
if (testStatType == 11) ropl.SetSubtractMLE(true);
ropl.SetPrintLevel(mPrintLevel);
ropl.SetMinimizer(mMinimizerType.c_str());
if (mEnableDetOutput) ropl.EnableDetailedOutput();
ProfileLikelihoodTestStat profll(*sbModel->GetPdf());
if (testStatType == 3) profll.SetOneSided(true);
if (testStatType == 4) profll.SetSigned(true);
profll.SetMinimizer(mMinimizerType.c_str());
profll.SetPrintLevel(mPrintLevel);
if (mEnableDetOutput) profll.EnableDetailedOutput();
profll.SetReuseNLL(mOptimize);
slrts.SetReuseNLL(mOptimize);
ropl.SetReuseNLL(mOptimize);
if (mOptimize) {
profll.SetStrategy(0);
ropl.SetStrategy(0);
}
if (mMaxPoi > 0) poi->
setMax(mMaxPoi);
MaxLikelihoodEstimateTestStat maxll(*sbModel->GetPdf(),*poi);
NumEventsTestStat nevtts;
AsymptoticCalculator::SetPrintLevel(mPrintLevel);
HypoTestCalculatorGeneric * hc = 0;
if (
type == 0) hc =
new FrequentistCalculator(*
data, *bModel, *sbModel);
else if (
type == 1) hc =
new HybridCalculator(*
data, *bModel, *sbModel);
else if (
type == 2 ) hc =
new AsymptoticCalculator(*
data, *bModel, *sbModel,
false );
else if (
type == 3 ) hc =
new AsymptoticCalculator(*
data, *bModel, *sbModel,
true );
else {
Error(
"StandardHypoTestInvDemo",
"Invalid - calculator type = %d supported values are only :\n\t\t\t 0 (Frequentist) , 1 (Hybrid) , 2 (Asymptotic) ",
type);
return 0;
}
TestStatistic * testStat = 0;
if (testStatType == 0) testStat = &slrts;
if (testStatType == 1 || testStatType == 11) testStat = &ropl;
if (testStatType == 2 || testStatType == 3 || testStatType == 4) testStat = &profll;
if (testStatType == 5) testStat = &maxll;
if (testStatType == 6) testStat = &nevtts;
if (testStat == 0) {
Error(
"StandardHypoTestInvDemo",
"Invalid - test statistic type = %d supported values are only :\n\t\t\t 0 (SLR) , 1 (Tevatron) , 2 (PLR), 3 (PLR1), 4(MLE)",testStatType);
return 0;
}
ToyMCSampler *toymcs = (ToyMCSampler*)hc->GetTestStatSampler();
if (toymcs && (
type == 0 ||
type == 1) ) {
if (sbModel->GetPdf()->canBeExtended() ) {
if (useNumberCounting)
Warning(
"StandardHypoTestInvDemo",
"Pdf is extended: but number counting flag is set: ignore it ");
}
else {
if (!useNumberCounting ) {
int nEvents =
data->numEntries();
Info(
"StandardHypoTestInvDemo",
"Pdf is not extended: number of events to generate taken from observed data set is %d",nEvents);
toymcs->SetNEventsPerToy(nEvents);
}
else {
Info(
"StandardHypoTestInvDemo",
"using a number counting pdf");
toymcs->SetNEventsPerToy(1);
}
}
toymcs->SetTestStatistic(testStat);
if (
data->isWeighted() && !mGenerateBinned) {
Info(
"StandardHypoTestInvDemo",
"Data set is weighted, nentries = %d and sum of weights = %8.1f but toy generation is unbinned - it would be faster to set mGenerateBinned to true\n",
data->numEntries(),
data->sumEntries());
}
toymcs->SetGenerateBinned(mGenerateBinned);
toymcs->SetUseMultiGen(mOptimize);
if (mGenerateBinned && sbModel->GetObservables()->getSize() > 2) {
Warning(
"StandardHypoTestInvDemo",
"generate binned is activated but the number of observable is %d. Too much memory could be needed for allocating all the bins",sbModel->GetObservables()->getSize() );
}
}
if (mReuseAltToys) {
hc->UseSameAltToys();
}
HybridCalculator *hhc = dynamic_cast<HybridCalculator*> (hc);
assert(hhc);
hhc->SetToys(ntoys,ntoys/mNToysRatio);
if (bModel->GetNuisanceParameters() || sbModel->GetNuisanceParameters() ) {
toymcs->SetUseMultiGen(false);
ToyMCSampler::SetAlwaysUseMultiGen(false);
if (nuisPriorName) nuisPdf = w->
pdf(nuisPriorName);
if (!nuisPdf) {
Info(
"StandardHypoTestInvDemo",
"No nuisance pdf given for the HybridCalculator - try to deduce pdf from the model");
if (bModel->GetPdf() && bModel->GetObservables() )
else
}
if (!nuisPdf ) {
if (bModel->GetPriorPdf()) {
nuisPdf = bModel->GetPriorPdf();
Info(
"StandardHypoTestInvDemo",
"No nuisance pdf given - try to use %s that is defined as a prior pdf in the B model",nuisPdf->
GetName());
}
else {
Error(
"StandardHypoTestInvDemo",
"Cannot run Hybrid calculator because no prior on the nuisance parameter is specified or can be derived");
return 0;
}
}
assert(nuisPdf);
Info(
"StandardHypoTestInvDemo",
"Using as nuisance Pdf ... " );
const RooArgSet * nuisParams = (bModel->GetNuisanceParameters() ) ? bModel->GetNuisanceParameters() : sbModel->GetNuisanceParameters();
Warning(
"StandardHypoTestInvDemo",
"Prior nuisance does not depend on nuisance parameters. They will be smeared in their full range");
}
delete np;
hhc->ForcePriorNuisanceAlt(*nuisPdf);
hhc->ForcePriorNuisanceNull(*nuisPdf);
}
}
if (testStatType == 3) ((AsymptoticCalculator*) hc)->SetOneSided(true);
if (testStatType != 2 && testStatType != 3)
Warning(
"StandardHypoTestInvDemo",
"Only the PL test statistic can be used with AsymptoticCalculator - use by default a two-sided PL");
}
((FrequentistCalculator*) hc)->SetToys(ntoys,ntoys/mNToysRatio);
if (mEnableDetOutput) ((FrequentistCalculator*) hc)->StoreFitInfo(true);
}
((HybridCalculator*) hc)->SetToys(ntoys,ntoys/mNToysRatio);
}
HypoTestInverter calc(*hc);
calc.SetConfidenceLevel(optHTInv.confLevel);
calc.UseCLs(useCLs);
calc.SetVerbose(true);
if (mUseProof) {
ProofConfig
pc(*w, mNWorkers,
"",
kFALSE);
toymcs->SetProofConfig(&
pc);
}
if (npoints > 0) {
if (poimin > poimax) {
poimin = int(poihat);
poimax = int(poihat + 4 * poi->
getError());
}
std::cout << "Doing a fixed scan in interval : " << poimin << " , " << poimax << std::endl;
calc.SetFixedScan(npoints,poimin,poimax);
}
else {
std::cout <<
"Doing an automatic scan in interval : " << poi->
getMin() <<
" , " << poi->
getMax() << std::endl;
}
HypoTestInverterResult *
r = calc.GetInterval();
std::cout << "Time to perform limit scan \n";
if (mRebuild) {
std::cout << "\n***************************************************************\n";
std::cout << "Rebuild the upper limit distribution by re-generating new set of pseudo-experiment and re-compute for each of them a new upper limit\n\n";
allParams = sbModel->GetPdf()->getParameters(*
data);
if (mRebuildParamValues != 0) {
*allParams = initialParameters;
}
if (mRebuildParamValues == 0 || mRebuildParamValues == 1 ) {
if (sbModel->GetNuisanceParameters() ) constrainParams.
add(*sbModel->GetNuisanceParameters());
const RooArgSet * poiModel = sbModel->GetParametersOfInterest();
bModel->LoadSnapshot();
if (mRebuildParamValues == 0 ) {
std::cout << "rebuild using fitted parameter value for B-model snapshot" << std::endl;
constrainParams.
Print(
"v");
}
}
std::cout << "StandardHypoTestInvDemo: Initial parameters used for rebuilding: ";
delete allParams;
calc.SetCloseProof(1);
SamplingDistribution * limDist = calc.GetUpperLimitDistribution(true,mNToyToRebuild);
std::cout << "Time to rebuild distributions " << std::endl;
if (limDist) {
std::cout << "Expected limits after rebuild distribution " << std::endl;
std::cout << "expected upper limit (median of limit distribution) " << limDist->InverseCDF(0.5) << std::endl;
std::cout <<
"expected -1 sig limit (0.16% quantile of limit dist) " << limDist->InverseCDF(
ROOT::Math::normal_cdf(-1)) << std::endl;
std::cout <<
"expected +1 sig limit (0.84% quantile of limit dist) " << limDist->InverseCDF(
ROOT::Math::normal_cdf(1)) << std::endl;
std::cout <<
"expected -2 sig limit (.025% quantile of limit dist) " << limDist->InverseCDF(
ROOT::Math::normal_cdf(-2)) << std::endl;
std::cout <<
"expected +2 sig limit (.975% quantile of limit dist) " << limDist->InverseCDF(
ROOT::Math::normal_cdf(2)) << std::endl;
SamplingDistPlot limPlot( (mNToyToRebuild < 200) ? 50 : 100);
limPlot.AddSamplingDistribution(limDist);
limPlot.GetTH1F()->SetStats(true);
limPlot.SetLineColor(
kBlue);
new TCanvas(
"limPlot",
"Upper Limit Distribution");
limPlot.Draw();
limDist->SetName("RULDist");
TFile * fileOut =
new TFile(
"RULDist.root",
"RECREATE");
limDist->Write();
}
else
std::cout << "ERROR : failed to re-build distributions " << std::endl;
}
}
void ReadResult(const char * fileName, const char * resultName="", bool useCLs=true) {
StandardHypoTestInvDemo(fileName, resultName,"","","",0,0,useCLs);
}
#ifdef USE_AS_MAIN
StandardHypoTestInvDemo();
}
#endif
void Info(const char *location, const char *msgfmt,...)
void Error(const char *location, const char *msgfmt,...)
void Warning(const char *location, const char *msgfmt,...)
R__EXTERN TSystem * gSystem
static void SetDefaultMinimizer(const char *type, const char *algo=0)
static void SetDefaultStrategy(int strat)
static const std::string & DefaultMinimizerType()
RooArgSet * getObservables(const RooArgSet &set, Bool_t valueOnly=kTRUE) const
virtual void Print(Option_t *options=0) const
Print TNamed name and title.
RooAbsCollection * snapshot(Bool_t deepCopy=kTRUE) const
Take a snap shot of current collection contents: An owning collection is returned containing clones o...
RooAbsArg * first() const
virtual void Print(Option_t *options=0) const
This method must be overridden when a class wants to print itself.
RooAbsData is the common abstract base class for binned and unbinned datasets.
static void setDefaultStorageType(StorageType s)
RooAbsPdf is the abstract interface for all probability density functions The class provides hybrid a...
virtual Double_t getMax(const char *name=0) const
virtual Double_t getMin(const char *name=0) const
Double_t getVal(const RooArgSet *set=0) const
Evaluate object. Returns either cached value or triggers a recalculation.
RooArgSet is a container object that can hold multiple RooAbsArg objects.
virtual Bool_t add(const RooAbsCollection &col, Bool_t silent=kFALSE)
Add a collection of arguments to this collection by calling add() for each element in the source coll...
RooFitResult is a container class to hold the input and output of a PDF fit to a dataset.
static RooMsgService & instance()
Return reference to singleton instance.
StreamConfig & getStream(Int_t id)
static TRandom * randomGenerator()
Return a pointer to a singleton random-number generator implementation.
RooRealVar represents a fundamental (non-derived) real valued object.
void setMax(const char *name, Double_t value)
Set maximum of name range to given value.
Double_t getError() const
virtual void setVal(Double_t value)
Set value of variable to 'value'.
The RooWorkspace is a persistable container for RooFit projects.
RooAbsData * data(const char *name) const
Retrieve dataset (binned or unbinned) with given name. A null pointer is returned if not found.
void Print(Option_t *opts=0) const
Print contents of the workspace.
TObject * obj(const char *name) const
Return any type of object (RooAbsArg, RooAbsData or generic object) with given name)
RooAbsPdf * pdf(const char *name) const
Retrieve p.d.f (RooAbsPdf) with given name. A null pointer is returned if not found.
virtual TObject * Get(const char *namecycle)
Return pointer to object identified by namecycle.
A ROOT file is a suite of consecutive data records (TKey instances) with a well defined format.
virtual void Close(Option_t *option="")
Close a file.
static TFile * Open(const char *name, Option_t *option="", const char *ftitle="", Int_t compress=ROOT::RCompressionSetting::EDefaults::kUseGeneralPurpose, Int_t netopt=0)
Create / open a file.
virtual const char * GetName() const
Returns name of object.
Mother of all ROOT objects.
virtual Int_t Write(const char *name=0, Int_t option=0, Int_t bufsize=0)
Write this object to the current directory.
virtual const char * GetName() const
Returns name of object.
virtual void SetSeed(ULong_t seed=0)
Set the random generator seed.
void Start(Bool_t reset=kTRUE)
Start the stopwatch.
void Print(Option_t *option="") const
Print the real and cpu time passed between the start and stop events.
static TString Format(const char *fmt,...)
Static method which formats a string using a printf style format descriptor and return a TString.
virtual Bool_t AccessPathName(const char *path, EAccessMode mode=kFileExists)
Returns FALSE if one can access a file using the specified access mode.
double normal_cdf(double x, double sigma=1, double x0=0)
Cumulative distribution function of the normal (Gaussian) distribution (lower tail).
int main(int argc, char **argv)
RooCmdArg Constrain(const RooArgSet ¶ms)
RooCmdArg Strategy(Int_t code)
RooCmdArg Hesse(Bool_t flag=kTRUE)
RooCmdArg InitialHesse(Bool_t flag=kTRUE)
RooCmdArg Save(Bool_t flag=kTRUE)
RooCmdArg PrintLevel(Int_t code)
RooCmdArg Offset(Bool_t flag=kTRUE)
RooCmdArg Minimizer(const char *type, const char *alg=0)
@(#)root/roostats:$Id$ Author: George Lewis, Kyle Cranmer
bool SetAllConstant(const RooAbsCollection &coll, bool constant=true)
void RemoveConstantParameters(RooArgSet *set)
RooAbsPdf * MakeNuisancePdf(RooAbsPdf &pdf, const RooArgSet &observables, const char *name)
void UseNLLOffset(bool on)
void PrintListContent(const RooArgList &l, std::ostream &os=std::cout)
static constexpr double pc
Double_t Sqrt(Double_t x)
Int_t CeilNint(Double_t x)
void removeTopic(RooFit::MsgTopic oldTopic)



 Standard tutorial macro for performing an inverted hypothesis test for computing an interval
Standard tutorial macro for performing an inverted hypothesis test for computing an interval