

Test program for the classes TUnfold and related.
Generate Monte Carlo and Data events The events consist of signal background
The signal is a resonance. It is generated with a Breit-Wigner, smeared by a Gaussian
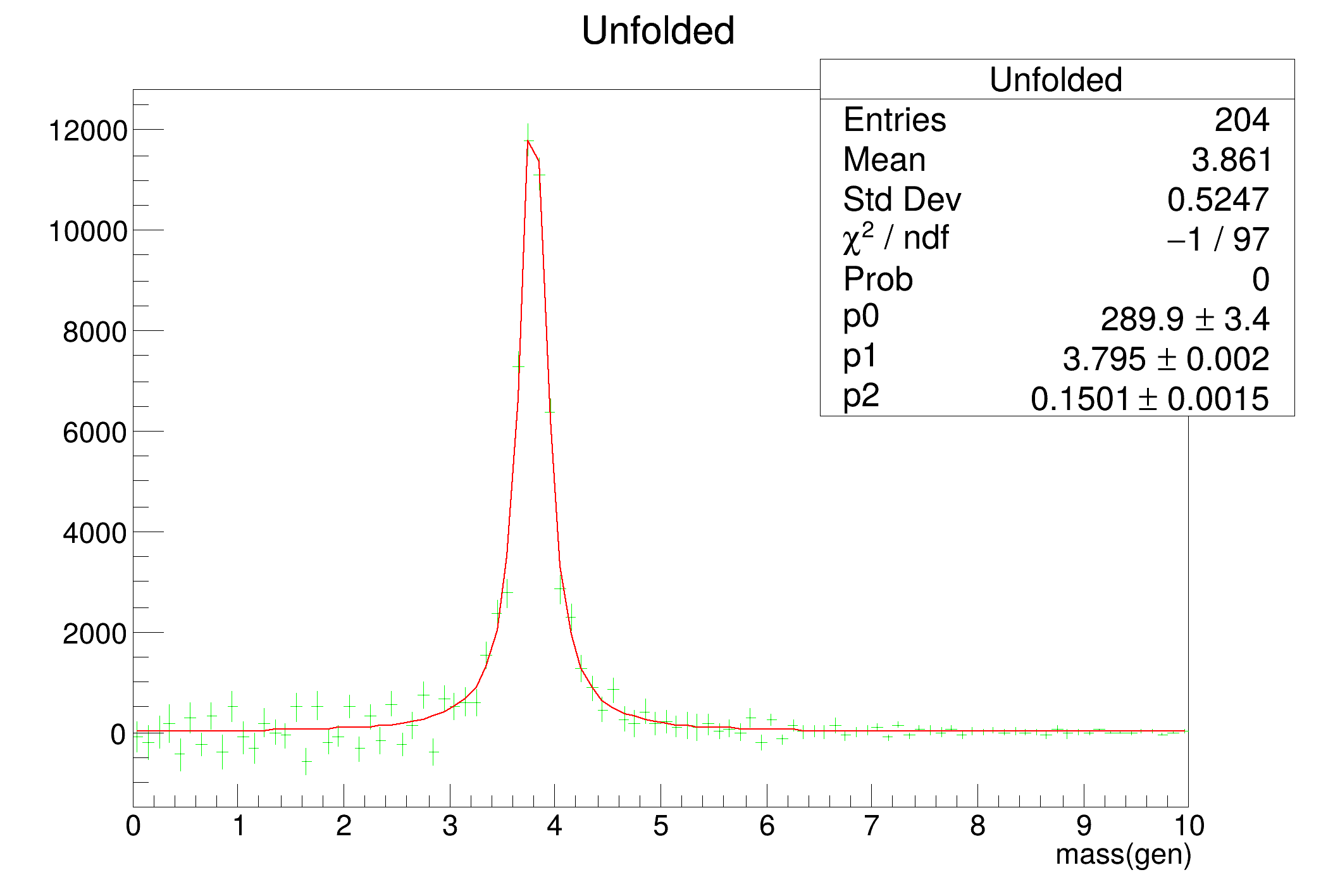
Unfold the data. The result is: The background level The shape of the resonance, corrected for detector effects
Systematic errors from the MC shape variation are included and propagated to the result
- fit the unfolded distribution, including the correlation matrix
- save six plots to a file
testUnfold1.ps
- 2d-plot of the matrix describing the migrations
- generator-level distributions
- blue: unfolded data, total errors
- green: unfolded data, statistical errors
- red: generated data
- black: fit to green data points
- detector level distributions
- blue: unfolded data, folded back through the matrix
- black: Monte Carlo (with wrong peal position)
- blue: data
- global correlation coefficients
- \( \chi^2 \) as a function of \( log(\tau) \) the star indicates the final choice of \( \tau \)
- the L curve
tau=5.56618e-05
chi**2=173.079+9.63005 / 147
chi**2(sys)=147.813
****************************************
Minimizer is Minuit2 / Migrad
MinFCN = 99.8432
NDf = 0
Edm = 2.41679e-07
NCalls = 110
p0 = 289.86 +/- 3.42424 -3.41527 +3.434 (Minos)
p1 = 3.79532 +/- 0.00174788 -0.00174785 +0.00174794 (Minos)
p2 = 0.15009 +/- 0.00146302 -0.00145983 +0.00146658 (Minos)
(int) 0
x =
hfit->GetBinCenter(i+1);
}
}
}
delete[] df;
}
return par[0]/(dm*dm+par[2]*par[2]);
}
{
do {
do {
} while(t>=1.0);
} while(t<=0.0);
return t;
} else {
do {
do {
} while(t>=1.0);
} while(t>=0.0);
return t;
}
}
} else {
}
}
{
4.0,
0.2);
}
(0.5,
3.6,
0.15);
}
3.8,
0.15);
}
}
std::cout<<"Unfolding result may be wrong\n";
}
#ifdef VERBOSE_LCURVE_SCAN
#endif
#ifdef VERBOSE_LCURVE_SCAN
#endif
}
}
}
std::cout<<
"tau="<<
unfold.GetTau()<<
"\n";
std::cout<<
"chi**2="<<
unfold.GetChi2A()<<
"+"<<
unfold.GetChi2L()
<<
" / "<<
unfold.GetNdf()<<
"\n";
std::cout<<
"chi**2(sys)="<<
unfold.GetChi2Sys()<<
"\n";
}
nullptr,
nullptr,
"*[UO]",
);
bw->SetParameter(0,1000.);
output.SaveAs(
"testUnfold1.ps");
return 0;
}
int Int_t
Signed integer 4 bytes (int)
double Double_t
Double 8 bytes.
ROOT::Detail::TRangeCast< T, true > TRangeDynCast
TRangeDynCast is an adapter class that allows the typed iteration through a TCollection.
Int_t gErrorIgnoreLevel
errors with level below this value will be ignored. Default is kUnset.
R__EXTERN TStyle * gStyle
static void RejectPoint(Bool_t reject=kTRUE)
Static function to set the global flag to reject points the fgRejectPoint global flag is tested by al...
virtual Int_t GetNpar() const
virtual void SetNumberFitPoints(Int_t npfits)
virtual void InitArgs(const Double_t *x, const Double_t *params)
Initialize parameters addresses.
virtual Double_t EvalPar(const Double_t *x, const Double_t *params=nullptr)
Evaluate function with given coordinates and parameters.
static Bool_t RejectedPoint()
See TF1::RejectPoint above.
A TGraph is an object made of two arrays X and Y with npoints each.
1-D histogram with a double per channel (see TH1 documentation)
TH1 is the base class of all histogram classes in ROOT.
static void SetDefaultSumw2(Bool_t sumw2=kTRUE)
When this static function is called with sumw2=kTRUE, all new histograms will automatically activate ...
2-D histogram with a double per channel (see TH1 documentation)
Service class for 2-D histogram classes.
Random number generator class based on M.
This is the base class for the ROOT Random number generators.
Base class for spline implementation containing the Draw/Paint methods.
void SetOptFit(Int_t fit=1)
The type of information about fit parameters printed in the histogram statistics box can be selected ...
An algorithm to unfold distributions from detector to truth level.
@ kSysErrModeRelative
matrix gives the relative shifts
@ kSysErrModeMatrix
matrix is an alternative to the default matrix, the errors are the difference to the original matrix
@ kHistMapOutputVert
truth level on y-axis of the response matrix
Abstract Base Class for Fitting.
static TVirtualFitter * Fitter(TObject *obj, Int_t maxpar=25)
Static function returning a pointer to the current fitter.
Double_t Sqrt(Double_t x)
Returns the square root of x.
LongDouble_t Power(LongDouble_t x, LongDouble_t y)
Returns x raised to the power y.
Double_t Tan(Double_t)
Returns the tangent of an angle of x radians.
Short_t Abs(Short_t d)
Returns the absolute value of parameter Short_t d.
Version 17.6, in parallel to changes in TUnfold
History:
- Version 17.5, in parallel to changes in TUnfold
- Version 17.4, in parallel to changes in TUnfold
- Version 17.3, in parallel to changes in TUnfold
- Version 17.2, in parallel to changes in TUnfold
- Version 17.1, in parallel to changes in TUnfold
- Version 17.0, updated for using the classes TUnfoldDensity, TUnfoldBinning
- Version 16.1, parallel to changes in TUnfold
- Version 16.0, parallel to changes in TUnfold
- Version 15, with automated L-curve scan
- Version 14, with changes in TUnfoldSys.cxx
- Version 13, include test of systematic errors
- Version 12, catch error when defining the input
- Version 11, print chi**2 and number of degrees of freedom
- Version 10, with bug-fix in TUnfold.cxx
- Version 9, with bug-fix in TUnfold.cxx and TUnfold.h
- Version 8, with bug-fix in TUnfold.cxx and TUnfold.h
- Version 7, with bug-fix in TUnfold.cxx and TUnfold.h
- Version 6a, fix problem with dynamic array allocation under windows
- Version 6, bug-fixes in TUnfold.C
- Version 5, replace main() by testUnfold1()
- Version 4, with bug-fix in TUnfold.C
- Version 3, with bug-fix in TUnfold.C
- Version 2, with changed ScanLcurve() arguments
- Version 1, remove L curve analysis, use ScanLcurve() method instead
- Version 0, L curve analysis included here
This file is part of TUnfold.
TUnfold is free software: you can redistribute it and/or modify it under the terms of the GNU General Public License as published by the Free Software Foundation, either version 3 of the License, or (at your option) any later version.
TUnfold is distributed in the hope that it will be useful, but WITHOUT ANY WARRANTY; without even the implied warranty of MERCHANTABILITY or FITNESS FOR A PARTICULAR PURPOSE. See the GNU General Public License for more details.
You should have received a copy of the GNU General Public License along with TUnfold. If not, see http://www.gnu.org/licenses/.
- Author
- Stefan Schmitt DESY, 14.10.2008
Definition in file testUnfold1.C.


