[#0] WARNING:InputArguments -- The parameter 'sigma1' with range [-inf, inf] of the RooGaussian 'sig1' exceeds the safe range of (0, inf). Advise to limit its range.
[#0] WARNING:InputArguments -- The parameter 'sigma2' with range [-inf, inf] of the RooGaussian 'sig2' exceeds the safe range of (0, inf). Advise to limit its range.
[#1] INFO:Minimization -- p.d.f. provides expected number of events, including extended term in likelihood.
[#1] INFO:Fitting -- RooAbsPdf::fitTo(model) fixing normalization set for coefficient determination to observables in data
[#1] INFO:Fitting -- using generic CPU library compiled with no vectorizations
[#1] INFO:Fitting -- Creation of NLL object took 875.233 μs
[#1] INFO:Fitting -- RooAddition::defaultErrorLevel(nll_model_modelData) Summation contains a RooNLLVar, using its error level
[#1] INFO:Minimization -- RooAbsMinimizerFcn::setOptimizeConst: activating const optimization
[#1] INFO:Minimization -- [fitFCN] No discrete parameters, performing continuous minimization only
[#1] INFO:Minimization -- RooAbsMinimizerFcn::setOptimizeConst: deactivating const optimization
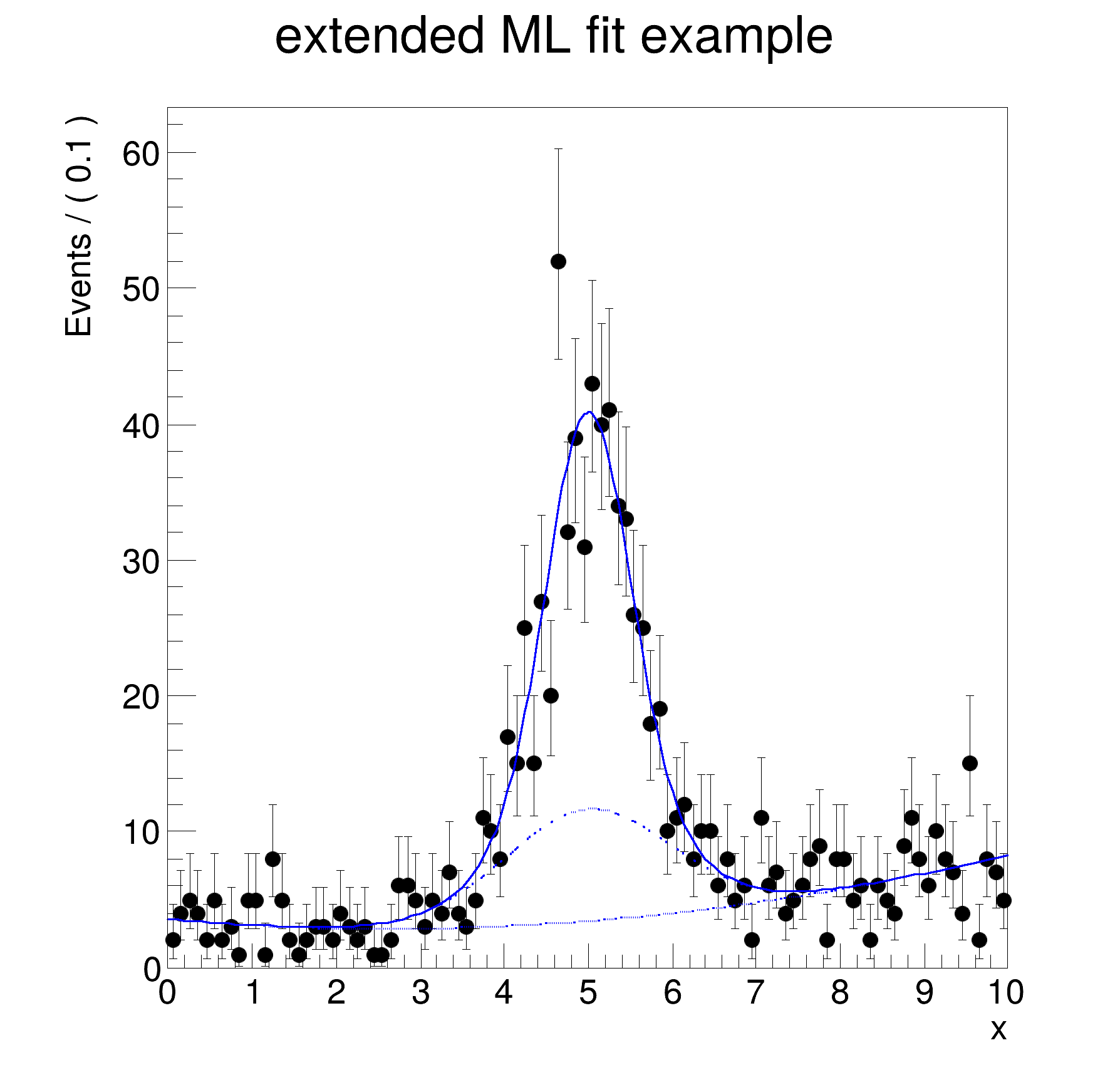
[#1] INFO:Plotting -- RooAbsPdf::plotOn(model) directly selected PDF components: (bkg)
[#1] INFO:Plotting -- RooAbsPdf::plotOn(model) indirectly selected PDF components: ()
[#1] INFO:Plotting -- RooAbsPdf::plotOn(model) directly selected PDF components: (bkg,sig2)
[#1] INFO:Plotting -- RooAbsPdf::plotOn(model) indirectly selected PDF components: (sig)
0x55e8db719de0 RooAddPdf::model = 0.885989/1 [Auto,Clean]
0x55e8db662400/V- RooChebychev::bkg = 0.733482 [Auto,Dirty]
0x55e8da45db60/V- RooRealVar::x = 5
0x55e8da66f2a0/V- RooRealVar::a0 = 0.507382 +/- 0.0795949
0x55e8da620490/V- RooRealVar::a1 = 0.266518 +/- 0.133887
0x55e8d6482580/V- RooRealVar::nbkg = 427.704 +/- 38.0643
0x55e8db718380/V- RooAddPdf::sig = 1/1 [Auto,Clean]
0x55e8d54f99d0/V- RooGaussian::sig1 = 1 [Auto,Dirty]
0x55e8da45db60/V- RooRealVar::x = 5
0x55e8da3c5680/V- RooRealVar::mean = 5
0x55e8da3af860/V- RooRealVar::sigma1 = 0.5
0x55e8daa176b0/V- RooRealVar::sig1frac = 0.640056 +/- 0.0966619
0x55e8da7af4a0/V- RooGaussian::sig2 = 1 [Auto,Dirty]
0x55e8da45db60/V- RooRealVar::x = 5
0x55e8da3c5680/V- RooRealVar::mean = 5
0x55e8d43c8a20/V- RooRealVar::sigma2 = 1
0x55e8daa1ce40/V- RooRealVar::nsig = 572.124 +/- 39.9094