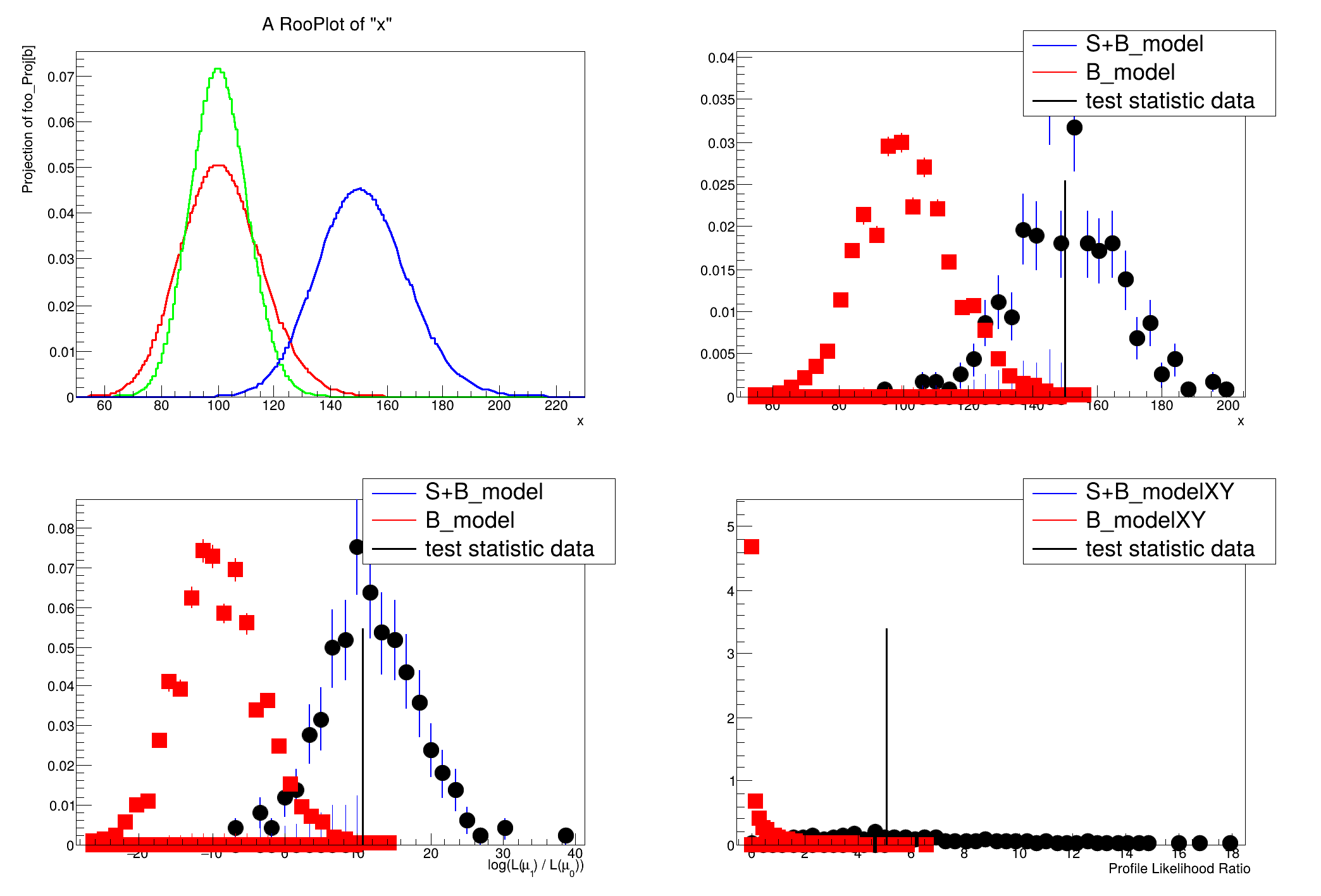
A hypothesis testing example based on number counting with background uncertainty.
NOTE: This example must be run with the ACLIC (the + option ) due to the new class that is defined.
The basic setup here is that a main measurement has observed x events with an expectation of s+b. One can choose an ad hoc prior for the uncertainty on b, or try to base it on an auxiliary measurement. In this case, the auxiliary measurement (aka control measurement, sideband) is another counting experiment with measurement y and expectation tau*b. With an 'original prior' on b, called \(\eta(b)\) then one can obtain a posterior from the auxiliary measurement \(\pi(b) = \eta(b) * Pois(y|tau*b).\) This is a principled choice for a prior on b in the main measurement of x, which can then be treated in a hybrid Bayesian/Frequentist way. Additionally, one can try to treat the two measurements simultaneously, which is detailed in Part 6 of the tutorial.
This tutorial is related to the FourBin.C tutorial in the modeling, but focuses on hypothesis testing instead of interval estimation.
More background on this 'prototype problem' can be found in the following papers:
[#1] INFO:NumericIntegration -- RooRealIntegral::init([py_X_prior_b_X_px]_Norm[b]_denominator_Int[b]) using numeric integrator RooIntegrator1D to calculate Int(b)
-----------------------------------------
Part 2
Hybrid p-value from direct integration = 0.000774094
Z_Gamma Significance = 3.1655
-----------------------------------------
Part 3
Z_Bi p-value (analytic): 0.00094165
Z_Bi significance (analytic): 3.10804
Real time 0:00:02, CP time 2.940
[#0] WARNING:Eval -- RooStatsUtils::MakeNuisancePdf - no constraints found on nuisance parameters in the input model
[#0] WARNING:Eval -- RooStatsUtils::MakeNuisancePdf - no constraints found on nuisance parameters in the input model
-----------------------------------------
Part 4
Results HypoTestCalculator_result:
- Null p-value = 0.0005 +/- 0.000288603
- Significance = 3.29053 +/- 0.162393 sigma
- Number of Alt toys: 300
- Number of Null toys: 6000
- Test statistic evaluated on data: 150
- CL_b: 0.0005 +/- 0.000288603
- CL_s+b: 0.506667 +/- 0.0288649
- CL_s: 1013.33 +/- 587.744
Real time 0:00:00, CP time 0.420
[#0] WARNING:Eval -- RooStatsUtils::MakeNuisancePdf - no constraints found on nuisance parameters in the input model
[#0] WARNING:Eval -- RooStatsUtils::MakeNuisancePdf - no constraints found on nuisance parameters in the input model
-----------------------------------------
Part 5
Results HypoTestCalculator_result:
- Null p-value = 0.000666667 +/- 0.000333222
- Significance = 3.20871 +/- 0.143724 sigma
- Number of Alt toys: 300
- Number of Null toys: 6000
- Test statistic evaluated on data: 10.8198
- CL_b: 0.000666667 +/- 0.000333222
- CL_s+b: 0.55 +/- 0.0287228
- CL_s: 825 +/- 414.607
Real time 0:00:00, CP time 0.540
[#0] WARNING:Eval -- RooStatsUtils::MakeNuisancePdf - no constraints found on nuisance parameters in the input model
[#0] WARNING:Eval -- RooStatsUtils::MakeNuisancePdf - no constraints found on nuisance parameters in the input model
-----------------------------------------
Part 6
Results HypoTestCalculator_result:
- Null p-value = 0.000666667 +/- 0.000333222
- Significance = 3.20871 +/- 0.143724 sigma
- Number of Alt toys: 300
- Number of Null toys: 6000
- Test statistic evaluated on data: 5.03388
- CL_b: 0.000666667 +/- 0.000333222
- CL_s+b: 0.493333 +/- 0.0288649
- CL_s: 740 +/- 372.402
Real time 0:00:03, CP time 3.200
public:
{
for (
int i = 0; i <
data.numEntries(); i++) {
value +=
data.get(i)->getRealValue(fColumnName.c_str());
}
}
virtual const TString GetVarName()
const {
return fColumnName; }
private:
string fColumnName;
protected:
};
{
w->factory(
"Poisson::px(x[150,0,500],sum::splusb(s[0,0,100],b[100,0.1,300]))");
w->factory(
"Poisson::py(y[100,0.1,500],prod::taub(tau[1.],b))");
w->factory(
"PROD::model(px,py)");
w->factory(
"Uniform::prior_b(b)");
w->factory(
"PROJ::averagedModel(PROD::foo(px|b,py,prior_b),b)");
w->var(
"s")->setVal(50.);
w->var(
"y")->setVal(100);
w->var(
"x")->setVal(150);
std::unique_ptr<RooAbsReal>
cdf{
w->pdf(
"averagedModel")->createCdf(*
w->var(
"x"))};
cout << "-----------------------------------------" << endl;
cout << "Part 2" << endl;
cout <<
"Hybrid p-value from direct integration = " << 1 -
cdf->getVal() << endl;
double p_Bi = NumberCountingUtils::BinomialWithTauObsP(150, 100, 1);
double Z_Bi = NumberCountingUtils::BinomialWithTauObsZ(150, 100, 1);
cout << "-----------------------------------------" << endl;
cout << "Part 3" << endl;
std::cout <<
"Z_Bi p-value (analytic): " <<
p_Bi << std::endl;
std::cout <<
"Z_Bi significance (analytic): " <<
Z_Bi << std::endl;
w->defineSet(
"obs",
"x");
w->defineSet(
"poi",
"s");
data->add(*
w->set(
"obs"));
b_model.SetParametersOfInterest(*
w->set(
"poi"));
w->var(
"s")->setVal(0.0);
sb_model.SetParametersOfInterest(*
w->set(
"poi"));
w->var(
"s")->setVal(50.0);
w->factory(
"Gaussian::gauss_prior(b,y, expr::sqrty('sqrt(y)',y))");
w->factory(
"Lognormal::lognorm_prior(b,y, expr::kappa('1+1./sqrt(y)',y))");
hc1.ForcePriorNuisanceAlt(*
w->pdf(
"py"));
hc1.ForcePriorNuisanceNull(*
w->pdf(
"py"));
cout << "-----------------------------------------" << endl;
cout << "Part 4" << endl;
hc2.ForcePriorNuisanceAlt(*
w->pdf(
"py"));
hc2.ForcePriorNuisanceNull(*
w->pdf(
"py"));
if (pc)
cout << "-----------------------------------------" << endl;
cout << "Part 5" << endl;
w->defineSet(
"obsXY",
"x,y");
w->var(
"x")->setVal(150.);
w->var(
"y")->setVal(100.);
b_modelXY.SetParametersOfInterest(*
w->set(
"poi"));
w->var(
"s")->setVal(0.0);
w->var(
"s")->setVal(50.0);
ropl.SetSubtractMLE(
false);
w->factory(
"Gamma::gamma_y0(b,sum::temp0(y0,1),1,0)");
w->factory(
"Gaussian::gauss_prior_y0(b,y0, expr::sqrty0('sqrt(y0)',y0))");
hc3.ForcePriorNuisanceAlt(*
w->pdf(
"gamma_y0"));
hc3.ForcePriorNuisanceNull(*
w->pdf(
"gamma_y0"));
if (pc)
cout << "-----------------------------------------" << endl;
cout << "Part 6" << endl;
}
#define ClassDef(name, id)
ROOT::Detail::TRangeCast< T, true > TRangeDynCast
TRangeDynCast is an adapter class that allows the typed iteration through a TCollection.
Option_t Option_t TPoint TPoint const char GetTextMagnitude GetFillStyle GetLineColor GetLineWidth GetMarkerStyle GetTextAlign GetTextColor GetTextSize void data
Option_t Option_t TPoint TPoint const char GetTextMagnitude GetFillStyle GetLineColor GetLineWidth GetMarkerStyle GetTextAlign GetTextColor GetTextSize void value
Abstract base class for binned and unbinned datasets.
RooArgSet is a container object that can hold multiple RooAbsArg objects.
Container class to hold unbinned data.
static RooMsgService & instance()
Return reference to singleton instance.
Plot frame and a container for graphics objects within that frame.
void Draw(Option_t *options=nullptr) override
Draw this plot and all of the elements it contains.
Same purpose as HybridCalculatorOriginal, but different implementation.
This class provides the plots for the result of a study performed with any of the HypoTestCalculatorG...
HypoTestResult is a base class for results from hypothesis tests.
MaxLikelihoodEstimateTestStat: TestStatistic that returns maximum likelihood estimate of a specified ...
ModelConfig is a simple class that holds configuration information specifying how a model should be u...
ProfileLikelihoodTestStat is an implementation of the TestStatistic interface that calculates the pro...
Holds configuration options for proof and proof-lite.
TestStatistic that returns the ratio of profiled likelihoods.
TestStatistic class that returns -log(L[null] / L[alt]) where L is the likelihood.
TestStatistic is an interface class to provide a facility for construction test statistics distributi...
ToyMCSampler is an implementation of the TestStatSampler interface.
Persistable container for RooFit projects.
void Divide(Int_t nx=1, Int_t ny=1, Float_t xmargin=0.01, Float_t ymargin=0.01, Int_t color=0) override
Automatic pad generation by division.
void Start(Bool_t reset=kTRUE)
Start the stopwatch.
void Stop()
Stop the stopwatch.
void Print(Option_t *option="") const override
Print the real and cpu time passed between the start and stop events.
RooCmdArg LineColor(Color_t color)
The namespace RooFit contains mostly switches that change the behaviour of functions of PDFs (or othe...
MsgLevel
Verbosity level for RooMsgService::StreamConfig in RooMsgService.
Namespace for the RooStats classes.
double PValueToSignificance(double pvalue)
returns one-sided significance corresponding to a p-value