import ROOT
ROOT.gStyle.SetOptStat(0)
ROOT.PyConfig.IgnoreCommandLineOptions = True
ROOT.gROOT.SetBatch(True)
observablename = "pTV"
obsvar = ROOT.RooRealVar(observablename, "observable of pTV", 10, 600)
kSM = ROOT.RooRealVar("kSM", "sm modifier", 1.0)
cHq3 = ROOT.RooRealVar("cHq3", "EFT modifier", -10.0, 10.0)
cHq3.setAttribute("NewPhysics", True)
cHl3 = ROOT.RooRealVar("cHl3", "EFT modifier", -10.0, 10.0)
cHl3.setAttribute("NewPhysics", True)
cHDD = ROOT.RooRealVar("cHDD", "EFT modifier", -10.0, 10.0)
cHDD.setAttribute("NewPhysics", True)
infilename = ROOT.gROOT.GetTutorialDir().Data() + "/roofit/input_histos_rf_lagrangianmorph.root"
par = "cHq3"
samplelist = [
"SM_NPsq0",
"cHq3_NPsq1",
"cHq3_NPsq2",
"cHl3_NPsq1",
"cHl3_NPsq2",
"cHDD_NPsq1",
"cHDD_NPsq2",
"cHl3_cHDD_NPsq2",
"cHq3_cHDD_NPsq2",
"cHl3_cHq3_NPsq2",
]
config = ROOT.RooLagrangianMorphFunc.Config()
config.fileName = infilename
config.observableName = observablename
config.folderNames = samplelist
config.couplings.add(cHq3)
config.couplings.add(cHDD)
config.couplings.add(cHl3)
config.couplings.add(kSM)
morphfunc = ROOT.RooLagrangianMorphFunc("morphfunc", "morphed dist. of pTV", config)
morphfunc.setParameter("cHq3", 0.01)
morphfunc.setParameter("cHl3", 1.0)
morphfunc.setParameter("cHDD", 0.2)
pseudo_hist = morphfunc.createTH1("pseudo_hist")
pseudo_dh = ROOT.RooDataHist("pseudo_dh", "pseudo_dh", [obsvar], pseudo_hist)
morphfunc.setParameter("cHq3", 0.0)
morphfunc.setParameter("cHl3", 0.0)
morphfunc.setParameter("cHDD", 0.0)
cHq3.setError(0.1)
cHl3.setError(0.1)
cHDD.setError(0.1)
model = ROOT.RooWrapperPdf("wrap_pdf", "wrap_pdf", morphfunc)
fitres = model.fitTo(pseudo_dh, SumW2Error=True, Optimize=False, Save=True, PrintLevel=-1)
hcorr = fitres.correlationHist()
postfit_hist = morphfunc.createTH1("morphing_postfit_hist")
postfit_dh = ROOT.RooDataHist("morphing_postfit_dh", "morphing_postfit_dh", [obsvar], postfit_hist)
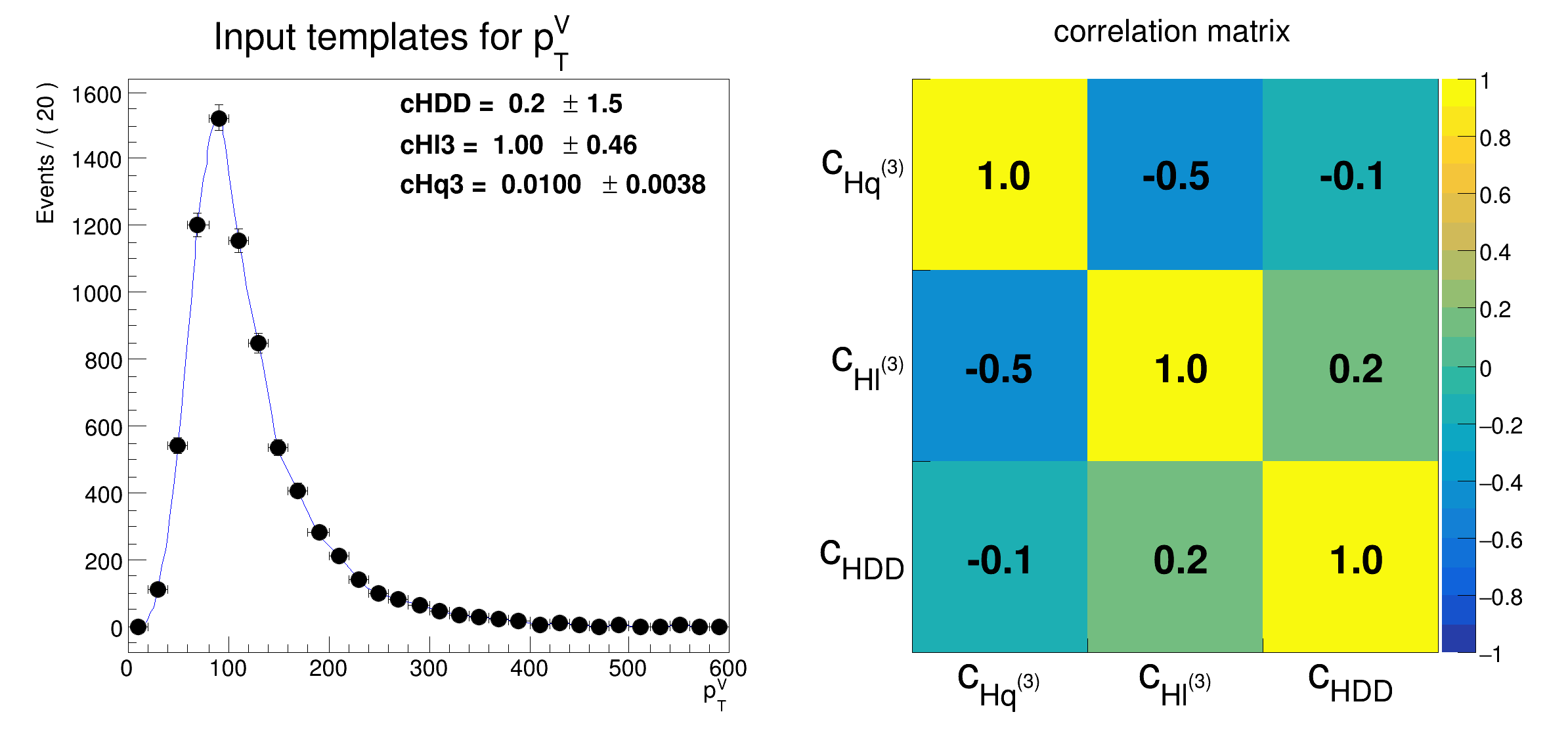
frame0 = obsvar.frame(Title="Input templates for p_{T}^{V}")
postfit_dh.plotOn(
frame0,
Name="postfit_dist",
DrawOption="C",
LineColor="b",
DataError=None,
XErrorSize=0,
)
pseudo_dh.plotOn(frame0, Name="input")
c1 = ROOT.TCanvas("fig3", "fig3", 800, 400)
c1.Divide(2, 1)
c1.cd(1)
ROOT.gPad.SetLeftMargin(0.15)
ROOT.gPad.SetRightMargin(0.05)
model.paramOn(frame0, ROOT.RooFit.Layout(0.50, 0.75, 0.9))
frame0.GetXaxis().SetTitle("p_{T}^{V}")
frame0.Draw()
c1.cd(2)
ROOT.gPad.SetLeftMargin(0.15)
ROOT.gPad.SetRightMargin(0.15)
ROOT.gStyle.SetPaintTextFormat("4.1f")
ROOT.gStyle.SetOptStat(0)
hcorr.SetMarkerSize(3.0)
hcorr.SetTitle("correlation matrix")
hcorr.GetYaxis().SetTitleOffset(1.4)
hcorr.GetYaxis().SetLabelSize(0.1)
hcorr.GetXaxis().SetLabelSize(0.1)
hcorr.GetYaxis().SetBinLabel(1, "c_{HDD}")
hcorr.GetYaxis().SetBinLabel(2, "c_{Hl^{(3)}}")
hcorr.GetYaxis().SetBinLabel(3, "c_{Hq^{(3)}}")
hcorr.GetXaxis().SetBinLabel(3, "c_{HDD}")
hcorr.GetXaxis().SetBinLabel(2, "c_{Hl^{(3)}}")
hcorr.GetXaxis().SetBinLabel(1, "c_{Hq^{(3)}}")
hcorr.GetYaxis().SetTitleOffset(1.4)
hcorr.Draw("colz text")
c1.SaveAs("rf712_lagrangianmorphfit.png")
[#0] PROGRESS:InputArguments -- initializing physics inputs from file /home/sftnight/build/workspace/root-makedoc-v630/rootspi/rdoc/src/v6-30-00-patches.build/tutorials/roofit/input_histos_rf_lagrangianmorph.root with object name(s) 'pTV'
[#0] PROGRESS:Caching -- creating cache from getCache function for 0x7a849e0
[#0] PROGRESS:Caching -- current storage has size 10
[#0] PROGRESS:ObjectHandling -- observable: pTV
[#0] PROGRESS:ObjectHandling -- binWidth: binWidth_pTV
[#1] INFO:DataHandling -- RooDataHist::adjustBinning(pseudo_dh): fit range of variable pTV expanded to nearest bin boundaries: [10,600] --> [0,600]
[#0] PROGRESS:Caching -- creating cache from getCache function for 0x9070460
[#0] PROGRESS:Caching -- current storage has size 10
[#0] PROGRESS:ObjectHandling -- observable: pTV
[#0] PROGRESS:ObjectHandling -- binWidth: binWidth_pTV
RooAbsMinimizerFcn: Minimized function has error status.
Returning maximum FCN so far (39620.3) to force MIGRAD to back out of this region. Error log follows.
Parameter values: cHDD=0.22238 cHl3=2.00088 cHq3=0.00780325
RooNLLVar::nll_wrap_pdf_pseudo_dh[ parameters=(binWidth_pTV,cHDD,cHl3,cHq3,kSM,nNP0,nNP1,nNP2,nNP3,nNP4) ]
function value is NAN @ parameters=(binWidth_pTV = 0.05,cHDD = 0.22238 +/- 0.1,cHl3 = 2.00088 +/- 0.1,cHq3 = 0.00780325 +/- 0.1,kSM = 1,nNP0 = 1,nNP1 = 1,nNP2 = 1,nNP3 = 1,nNP4 = 1)
RooWrapperPdf::wrap_pdf[ inputFunction=morphfunc ]
p.d.f value is less than zero (-0.040406), trying to recover @ inputFunction=morphfunc=-0.0404061
getLogVal() top-level p.d.f evaluates to NaN @ inputFunction=morphfunc=-0.0404061
p.d.f value is less than zero (-0.040462), trying to recover @ inputFunction=morphfunc=-0.0404616
getLogVal() top-level p.d.f evaluates to NaN @ inputFunction=morphfunc=-0.0404616
RooAbsMinimizerFcn: Minimized function has error status.
Returning maximum FCN so far (39620.3) to force MIGRAD to back out of this region. Error log follows.
Parameter values: cHDD=1.16153 cHl3=8.6965 cHq3=0.0408466
RooNLLVar::nll_wrap_pdf_pseudo_dh[ parameters=(binWidth_pTV,cHDD,cHl3,cHq3,kSM,nNP0,nNP1,nNP2,nNP3,nNP4) ]
function value is NAN @ parameters=(binWidth_pTV = 0.05,cHDD = 1.16153 +/- 0.1,cHl3 = 8.6965 +/- 0.1,cHq3 = 0.0408466 +/- 0.1,kSM = 1,nNP0 = 1,nNP1 = 1,nNP2 = 1,nNP3 = 1,nNP4 = 1)
RooWrapperPdf::wrap_pdf[ inputFunction=morphfunc ]
p.d.f value is less than zero (-0.423644), trying to recover @ inputFunction=morphfunc=-0.423644
getLogVal() top-level p.d.f evaluates to NaN @ inputFunction=morphfunc=-0.423644
p.d.f value is less than zero (-2.779762), trying to recover @ inputFunction=morphfunc=-2.77976
getLogVal() top-level p.d.f evaluates to NaN @ inputFunction=morphfunc=-2.77976
p.d.f value is less than zero (-0.022566), trying to recover @ inputFunction=morphfunc=-0.0225659
getLogVal() top-level p.d.f evaluates to NaN @ inputFunction=morphfunc=-0.0225659
p.d.f value is less than zero (-0.205861), trying to recover @ inputFunction=morphfunc=-0.205861
getLogVal() top-level p.d.f evaluates to NaN @ inputFunction=morphfunc=-0.205861
p.d.f value is less than zero (-1.337222), trying to recover @ inputFunction=morphfunc=-1.33722
getLogVal() top-level p.d.f evaluates to NaN @ inputFunction=morphfunc=-1.33722
p.d.f value is less than zero (-0.477718), trying to recover @ inputFunction=morphfunc=-0.477718
getLogVal() top-level p.d.f evaluates to NaN @ inputFunction=morphfunc=-0.477718
... (remaining 4 messages suppressed)
RooAbsMinimizerFcn: Minimized function has error status.
Returning maximum FCN so far (39620.3) to force MIGRAD to back out of this region. Error log follows.
Parameter values: cHDD=0.607791 cHl3=5.23432 cHq3=0.0213386
RooNLLVar::nll_wrap_pdf_pseudo_dh[ parameters=(binWidth_pTV,cHDD,cHl3,cHq3,kSM,nNP0,nNP1,nNP2,nNP3,nNP4) ]
function value is NAN @ parameters=(binWidth_pTV = 0.05,cHDD = 0.607791 +/- 0.1,cHl3 = 5.23432 +/- 0.1,cHq3 = 0.0213386 +/- 0.1,kSM = 1,nNP0 = 1,nNP1 = 1,nNP2 = 1,nNP3 = 1,nNP4 = 1)
RooWrapperPdf::wrap_pdf[ inputFunction=morphfunc ]
p.d.f value is less than zero (-0.067384), trying to recover @ inputFunction=morphfunc=-0.0673836
getLogVal() top-level p.d.f evaluates to NaN @ inputFunction=morphfunc=-0.0673836
p.d.f value is less than zero (-0.313216), trying to recover @ inputFunction=morphfunc=-0.313216
getLogVal() top-level p.d.f evaluates to NaN @ inputFunction=morphfunc=-0.313216
p.d.f value is less than zero (-1.542083), trying to recover @ inputFunction=morphfunc=-1.54208
getLogVal() top-level p.d.f evaluates to NaN @ inputFunction=morphfunc=-1.54208
p.d.f value is less than zero (-1.460439), trying to recover @ inputFunction=morphfunc=-1.46044
getLogVal() top-level p.d.f evaluates to NaN @ inputFunction=morphfunc=-1.46044
p.d.f value is less than zero (-1.443873), trying to recover @ inputFunction=morphfunc=-1.44387
getLogVal() top-level p.d.f evaluates to NaN @ inputFunction=morphfunc=-1.44387
p.d.f value is less than zero (-1.909523), trying to recover @ inputFunction=morphfunc=-1.90952
getLogVal() top-level p.d.f evaluates to NaN @ inputFunction=morphfunc=-1.90952
... (remaining 16 messages suppressed)
RooAbsMinimizerFcn: Minimized function has error status.
Returning maximum FCN so far (39620.3) to force MIGRAD to back out of this region. Error log follows.
Parameter values: cHDD=0.330108 cHl3=2.94627 cHq3=0.0115846
RooNLLVar::nll_wrap_pdf_pseudo_dh[ parameters=(binWidth_pTV,cHDD,cHl3,cHq3,kSM,nNP0,nNP1,nNP2,nNP3,nNP4) ]
function value is NAN @ parameters=(binWidth_pTV = 0.05,cHDD = 0.330108 +/- 0.1,cHl3 = 2.94627 +/- 0.1,cHq3 = 0.0115846 +/- 0.1,kSM = 1,nNP0 = 1,nNP1 = 1,nNP2 = 1,nNP3 = 1,nNP4 = 1)
RooWrapperPdf::wrap_pdf[ inputFunction=morphfunc ]
p.d.f value is less than zero (-0.072286), trying to recover @ inputFunction=morphfunc=-0.0722861
getLogVal() top-level p.d.f evaluates to NaN @ inputFunction=morphfunc=-0.0722861
p.d.f value is less than zero (-0.082234), trying to recover @ inputFunction=morphfunc=-0.0822337
getLogVal() top-level p.d.f evaluates to NaN @ inputFunction=morphfunc=-0.0822337
p.d.f value is less than zero (-0.149372), trying to recover @ inputFunction=morphfunc=-0.149372
getLogVal() top-level p.d.f evaluates to NaN @ inputFunction=morphfunc=-0.149372
RooAbsMinimizerFcn: Minimized function has error status.
Returning maximum FCN so far (39620.3) to force MIGRAD to back out of this region. Error log follows.
Parameter values: cHDD=0.191157 cHl3=1.72296 cHq3=0.00670752
RooNLLVar::nll_wrap_pdf_pseudo_dh[ parameters=(binWidth_pTV,cHDD,cHl3,cHq3,kSM,nNP0,nNP1,nNP2,nNP3,nNP4) ]
function value is NAN @ parameters=(binWidth_pTV = 0.05,cHDD = 0.191157 +/- 0.1,cHl3 = 1.72296 +/- 0.1,cHq3 = 0.00670752 +/- 0.1,kSM = 1,nNP0 = 1,nNP1 = 1,nNP2 = 1,nNP3 = 1,nNP4 = 1)
RooWrapperPdf::wrap_pdf[ inputFunction=morphfunc ]
p.d.f value is less than zero (-0.027317), trying to recover @ inputFunction=morphfunc=-0.0273169
getLogVal() top-level p.d.f evaluates to NaN @ inputFunction=morphfunc=-0.0273169
p.d.f value is less than zero (-0.007139), trying to recover @ inputFunction=morphfunc=-0.00713888
getLogVal() top-level p.d.f evaluates to NaN @ inputFunction=morphfunc=-0.00713888
RooAbsMinimizerFcn: Minimized function has error status.
Returning maximum FCN so far (39620.3) to force MIGRAD to back out of this region. Error log follows.
Parameter values: cHDD=0.138238 cHl3=1.24892 cHq3=0.00485049
RooNLLVar::nll_wrap_pdf_pseudo_dh[ parameters=(binWidth_pTV,cHDD,cHl3,cHq3,kSM,nNP0,nNP1,nNP2,nNP3,nNP4) ]
function value is NAN @ parameters=(binWidth_pTV = 0.05,cHDD = 0.138238 +/- 0.1,cHl3 = 1.24892 +/- 0.1,cHq3 = 0.00485049 +/- 0.1,kSM = 1,nNP0 = 1,nNP1 = 1,nNP2 = 1,nNP3 = 1,nNP4 = 1)
RooWrapperPdf::wrap_pdf[ inputFunction=morphfunc ]
p.d.f value is less than zero (-0.001109), trying to recover @ inputFunction=morphfunc=-0.00110901
getLogVal() top-level p.d.f evaluates to NaN @ inputFunction=morphfunc=-0.00110901
RooAbsMinimizerFcn: Minimized function has error status.
Returning maximum FCN so far (39620.3) to force MIGRAD to back out of this region. Error log follows.
Parameter values: cHDD=0.121667 cHl3=1.24299 cHq3=0.004269
RooNLLVar::nll_wrap_pdf_pseudo_dh[ parameters=(binWidth_pTV,cHDD,cHl3,cHq3,kSM,nNP0,nNP1,nNP2,nNP3,nNP4) ]
function value is NAN @ parameters=(binWidth_pTV = 0.05,cHDD = 0.121667 +/- 0.1,cHl3 = 1.24299 +/- 0.1,cHq3 = 0.004269 +/- 0.1,kSM = 1,nNP0 = 1,nNP1 = 1,nNP2 = 1,nNP3 = 1,nNP4 = 1)
RooWrapperPdf::wrap_pdf[ inputFunction=morphfunc ]
p.d.f value is less than zero (-0.001164), trying to recover @ inputFunction=morphfunc=-0.00116395
getLogVal() top-level p.d.f evaluates to NaN @ inputFunction=morphfunc=-0.00116395
[#1] INFO:Fitting -- RooAbsPdf::fitTo(wrap_pdf) Calculating sum-of-weights-squared correction matrix for covariance matrix
[#1] INFO:InputArguments -- RooAbsData::plotOn(pseudo_dh) INFO: dataset has non-integer weights, auto-selecting SumW2 errors instead of Poisson errors
[#1] INFO:Plotting -- RooPlot::updateFitRangeNorm: New event count of 7389.24 will supersede previous event count of 7388.38 for normalization of PDF projections