

Multidimensional models: marginizalization of multi-dimensional pdfs through integration
import ROOT
ROOT.RooAbsPdf.defaultIntegratorConfig().setEpsRel(1e-8)
ROOT.RooAbsPdf.defaultIntegratorConfig().setEpsAbs(1e-8)
x = ROOT.RooRealVar("x", "x", -5, 5)
y = ROOT.RooRealVar("y", "y", -2, 2)
a0 = ROOT.RooRealVar("a0", "a0", 0)
a1 = ROOT.RooRealVar("a1", "a1", -1.5, -3, 1)
fy = ROOT.RooPolyVar("fy", "fy", y, [a0, a1])
sigmax = ROOT.RooRealVar("sigmax", "width of gaussian", 0.5)
gaussx = ROOT.RooGaussian("gaussx", "Gaussian in x with shifting mean in y", x, fy, sigmax)
gaussy = ROOT.RooGaussian("gaussy", "Gaussian in y", y, 0.0, 2.0)
model = ROOT.RooProdPdf(
"model",
"gaussx(x|y)*gaussy(y)",
{gaussy},
Conditional=({gaussx}, {x}),
)
modelx = model.createProjection({y})
data = modelx.generateBinned({x}, 1000)
modelx.fitTo(data, Verbose=True, PrintLevel=-1)
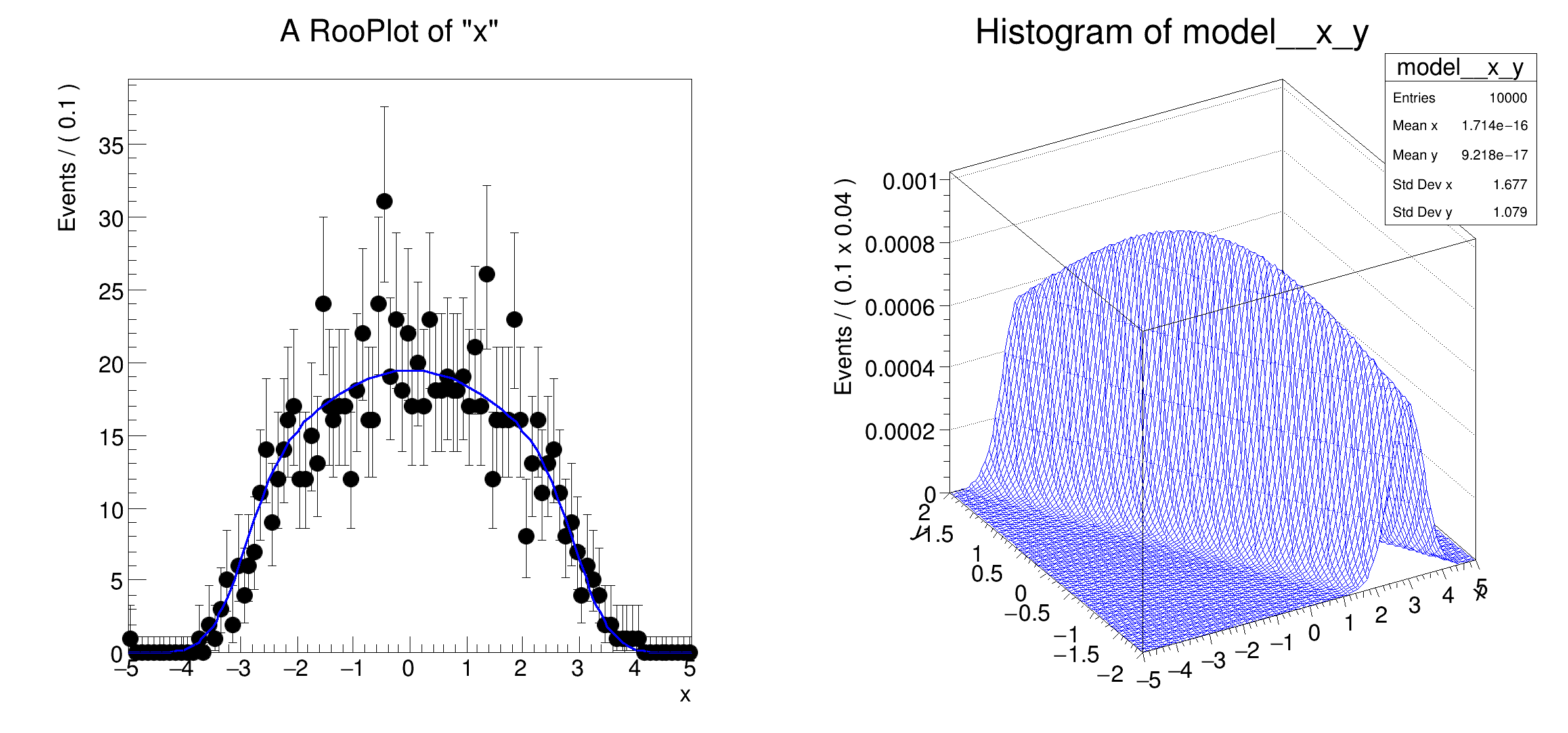
frame = x.frame(40)
data.plotOn(frame)
modelx.plotOn(frame)
hh = model.createHistogram("x,y")
hh.SetLineColor(ROOT.kBlue)
c = ROOT.TCanvas("rf315_projectpdf", "rf315_projectpdf", 800, 400)
c.Divide(2)
c.cd(1)
ROOT.gPad.SetLeftMargin(0.15)
frame.GetYaxis().SetTitleOffset(1.4)
frame.Draw()
c.cd(2)
ROOT.gPad.SetLeftMargin(0.20)
hh.GetZaxis().SetTitleOffset(2.5)
hh.Draw("surf")
c.SaveAs("rf315_projectpdf.png")
[#0] WARNING:InputArguments -- The parameter 'sigmax' with range [-inf, inf] of the RooGaussian 'gaussx' exceeds the safe range of (0, inf). Advise to limit its range.
[#1] INFO:NumericIntegration -- RooRealIntegral::init([gaussy_NORM[y]_X_gaussx_NORM[x]]_Int[y]) using numeric integrator RooRombergIntegrator to calculate Int(y)
[#1] INFO:NumericIntegration -- RooRealIntegral::init([gaussy_NORM[y]_X_gaussx_NORM[x]]_Int[y]) using numeric integrator RooRombergIntegrator to calculate Int(y)
[#1] INFO:Minimization -- RooAbsMinimizerFcn::setOptimizeConst: activating const optimization
[#1] INFO:Minimization -- The following expressions will be evaluated in cache-and-track mode: (gaussx)
[#0] WARNING:Minimization -- RooAbsMinimizerFcn::synchronize: WARNING: no initial error estimate available for a1: using 0.4
prevFCN = 1900.156536 a1=-1.488,
prevFCN = 1899.96972 a1=-1.512,
prevFCN = 1900.566064 a1=-1.499,
prevFCN = 1900.127678 a1=-1.501,
prevFCN = 1900.187622 a1=-1.484,
prevFCN = 1899.958651 a1=-1.483,
prevFCN = 1899.959674 a1=-1.484,
prevFCN = 1899.958586 a1=-1.484,
prevFCN = 1899.958651 a1=-1.483,
prevFCN = 1899.959674 a1=-1.484,
prevFCN = 1899.958586 a1=-1.483,
prevFCN = 1899.958779 a1=-1.484,
prevFCN = 1899.958562 a1=-1.484,
prevFCN = 1899.958651 a1=-1.483,
prevFCN = 1899.958779 a1=-1.484,
prevFCN = 1899.958562 a1=-1.484,
prevFCN = 1899.958674 a1=-1.484,
prevFCN = 1899.95863 a1=-1.484, [#1] INFO:Minimization -- RooAbsMinimizerFcn::setOptimizeConst: deactivating const optimization
[#1] INFO:NumericIntegration -- RooRealIntegral::init([gaussy_NORM[y]_X_gaussx_NORM[x]]_Int[y]) using numeric integrator RooRombergIntegrator to calculate Int(y)
- Date
- February 2018
- Authors
- Clemens Lange, Wouter Verkerke (C++ version)
Definition in file rf315_projectpdf.py.


