import ROOT
import numpy as np
iparB = np.array([0, 2], dtype=np.int32)
iparSB = np.array(
[
1,
2,
3,
4,
5,
],
dtype=np.int32,
)
def __init__(self, f1, f2):
self._f1 = f1
self._f2 = f2
def __call__(self, par):
par_arr = np.frombuffer(par, dtype=np.float64, count=6)
p1 = par_arr[iparB]
p2 = par_arr[iparSB]
return self._f1(p1) + self._f2(p2)
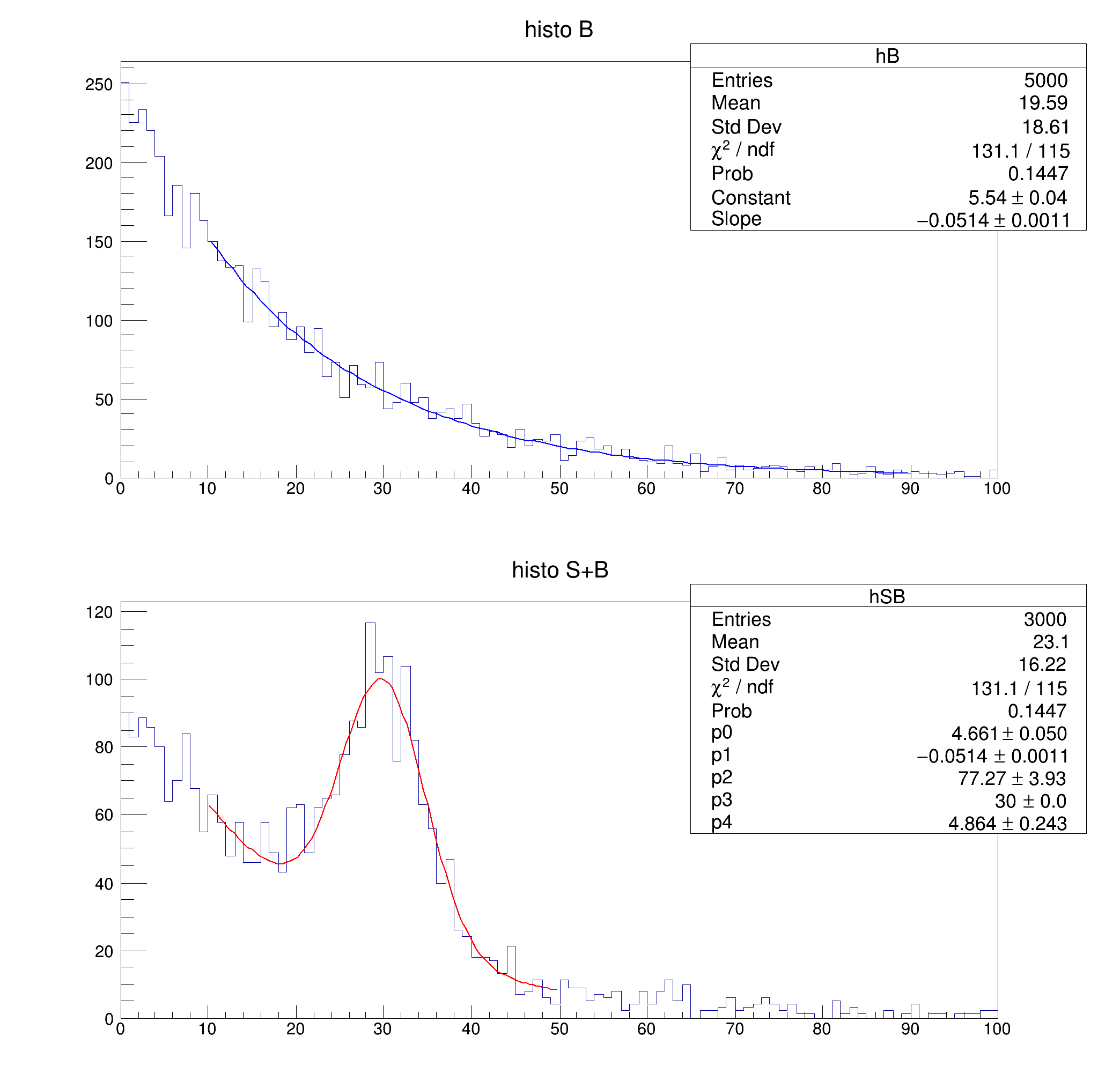
hB = ROOT.TH1D("hB", "histo B", 100, 0, 100)
hSB = ROOT.TH1D("hSB", "histo S+B", 100, 0, 100)
fB = ROOT.TF1("fB", "expo", 0, 100)
fB.SetParameters(1, -0.05)
hB.FillRandom("fB")
fS = ROOT.TF1("fS", "gaus", 0, 100)
fS.SetParameters(1, 30, 5)
hSB.FillRandom("fB", 2000)
hSB.FillRandom("fS", 1000)
fSB = ROOT.TF1("fSB", "expo + gaus(2)", 0, 100)
rangeB.SetRange(10, 90)
rangeSB.SetRange(10, 50)
globalChi2 = GlobalChi2(chi2_B, chi2_SB)
Npar = 6
par0 = np.array([5, 5, -0.1, 100, 30, 10])
fitter.Config().SetParamsSettings(6, par0)
fitter.Config().ParSettings(4).Fix()
fitter.Config().ParSettings(2).SetLimits(-10, -1.0e-4)
fitter.Config().ParSettings(3).SetLimits(0, 10000)
fitter.Config().ParSettings(3).SetStepSize(5)
fitter.Config().MinimizerOptions().SetPrintLevel(0)
fitter.Config().SetMinimizer("Minuit2", "Migrad")
fitter.FitFCN(globalChi2Functor, 0, dataB.Size() + dataSB.Size(), True)
result = fitter.Result()
result.Print(ROOT.std.cout)
c1 = ROOT.TCanvas("Simfit", "Simultaneous fit of two histograms", 10, 10, 700, 700)
c1.Divide(1, 2)
c1.cd(1)
ROOT.gStyle.SetOptFit(1111)
fB.SetFitResult(result, iparB)
fB.SetRange(rangeB().first, rangeB().second)
fB.SetLineColor(ROOT.kBlue)
hB.GetListOfFunctions().
Add(fB)
hB.Draw()
c1.cd(2)
fSB.SetFitResult(result, iparSB)
fSB.SetRange(rangeSB().first, rangeSB().second)
fSB.SetLineColor(ROOT.kRed)
hSB.GetListOfFunctions().
Add(fSB)
hSB.Draw()
c1.SaveAs("combinedFit.png")
Class describing the binned data sets : vectors of x coordinates, y values and optionally error on y ...
Chi2FCN class for binned fits using the least square methods.
class describing the range in the coordinates it supports multiple range in a coordinate.
Fitter class, entry point for performing all type of fits.
Documentation for class Functor class.
Class to Wrap a ROOT Function class (like TF1) in a IParamMultiFunction interface of multi-dimensions...
void FillData(BinData &dv, const TH1 *hist, TF1 *func=nullptr)
fill the data vector from a TH1.
TMatrixT< Element > & Add(TMatrixT< Element > &target, Element scalar, const TMatrixT< Element > &source)
Modify addition: target += scalar * source.
DataOptions : simple structure holding the options on how the data are filled.