␛[1mRooFit v3.60 -- Developed by Wouter Verkerke and David Kirkby␛[0m
Copyright (C) 2000-2013 NIKHEF, University of California & Stanford University
All rights reserved, please read http://roofit.sourceforge.net/license.txt
[#0] PROGRESS:InputArguments -- initializing physics inputs from file /home/sftnight/build/workspace/root-makedoc-v626/rootspi/rdoc/src/v6-26-00-patches.build/tutorials/roofit/input_histos_rf_lagrangianmorph.root with object name(s) 'pTV'
[#0] PROGRESS:Caching -- creating cache from getCache function for 0x7fffb8aa6f98
[#0] PROGRESS:Caching -- current storage has size 3
[#0] PROGRESS:ObjectHandling -- observable: pTV
[#0] PROGRESS:ObjectHandling -- binWidth: binWidth_pTV
[#1] INFO:DataHandling -- RooDataHist::adjustBinning(morph_dh_cHq3=0.01): fit range of variable pTV expanded to nearest bin boundaries: [10,600] --> [0,600]
[#1] INFO:DataHandling -- RooDataHist::adjustBinning(morph_dh_cHq3=0.25): fit range of variable pTV expanded to nearest bin boundaries: [0,600] --> [0,600]
[#1] INFO:DataHandling -- RooDataHist::adjustBinning(morph_dh_cHq3=0.5): fit range of variable pTV expanded to nearest bin boundaries: [0,600] --> [0,600]
[#1] INFO:InputArguments -- RooAbsData::plotOn(SM_NPsq0) INFO: dataset has non-integer weights, auto-selecting SumW2 errors instead of Poisson errors
[#1] INFO:InputArguments -- RooAbsData::plotOn(cHq3_NPsq1) INFO: dataset has non-integer weights, auto-selecting SumW2 errors instead of Poisson errors
[#1] INFO:Plotting -- RooPlot::updateFitRangeNorm: New event count of 24931.9 will supercede previous event count of 10852.3 for normalization of PDF projections
[#1] INFO:InputArguments -- RooAbsData::plotOn(cHq3_NPsq2) INFO: dataset has non-integer weights, auto-selecting SumW2 errors instead of Poisson errors
[#1] INFO:Plotting -- RooPlot::updateFitRangeNorm: New event count of 29789.2 will supercede previous event count of 24931.9 for normalization of PDF projections
[#1] INFO:Plotting -- RooPlot::updateFitRangeNorm: New event count of 18947.1 will supercede previous event count of 11104.6 for normalization of PDF projections
[#1] INFO:Plotting -- RooPlot::updateFitRangeNorm: New event count of 30765.5 will supercede previous event count of 18947.1 for normalization of PDF projections
[#0] PROGRESS:Caching -- creating cache from getCache function for 0x55894493f0e0
[#0] PROGRESS:Caching -- current storage has size 3
[#0] PROGRESS:ObjectHandling -- observable: pTV
[#0] PROGRESS:ObjectHandling -- binWidth: binWidth_pTV
[#1] INFO:NumericIntegration -- RooRealIntegral::init(w_SM_NPsq0_morphfunc_Int[cHq3]) using numeric integrator RooIntegrator1D to calculate Int(cHq3)
[#1] INFO:NumericIntegration -- RooRealIntegral::init(w_cHq3_NPsq1_morphfunc_Int[cHq3]) using numeric integrator RooIntegrator1D to calculate Int(cHq3)
[#1] INFO:NumericIntegration -- RooRealIntegral::init(w_cHq3_NPsq2_morphfunc_Int[cHq3]) using numeric integrator RooIntegrator1D to calculate Int(cHq3)
[#0] PROGRESS:Caching -- creating cache from getCache function for 0x558944eefbb0
[#0] PROGRESS:Caching -- current storage has size 3
[#0] PROGRESS:ObjectHandling -- observable: pTV
[#0] PROGRESS:ObjectHandling -- binWidth: binWidth_pTV
[#1] INFO:NumericIntegration -- RooRealIntegral::init(w_SM_NPsq0_morphfunc_Int[cHq3]) using numeric integrator RooIntegrator1D to calculate Int(cHq3)
[#1] INFO:NumericIntegration -- RooRealIntegral::init(w_cHq3_NPsq1_morphfunc_Int[cHq3]) using numeric integrator RooIntegrator1D to calculate Int(cHq3)
[#1] INFO:NumericIntegration -- RooRealIntegral::init(w_cHq3_NPsq2_morphfunc_Int[cHq3]) using numeric integrator RooIntegrator1D to calculate Int(cHq3)
{
std::string observablename = "pTV";
RooRealVar obsvar(observablename.c_str(),
"p_{T}^{V}", 10, 600);
RooRealVar cHq3(
"cHq3",
"EFT modifier", 0.0, 1.0);
cHq3.setAttribute("NewPhysics", true);
std::string infilename = std::string(
gROOT->GetTutorialDir()) +
"/roofit/input_histos_rf_lagrangianmorph.root";
std::vector<std::string> samplelist = {"SM_NPsq0", "cHq3_NPsq1", "cHq3_NPsq2"};
morphfunc.setParameter("cHq3", 0.01);
auto morph_hist_0p01 = morphfunc.createTH1("morph_cHq3=0.01");
morphfunc.setParameter("cHq3", 0.25);
auto morph_hist_0p25 = morphfunc.createTH1("morph_cHq3=0.25");
morphfunc.setParameter("cHq3", 0.5);
auto morph_hist_0p5 = morphfunc.createTH1("morph_cHq3=0.5");
file->GetObject(samplelist[0].c_str(), folder);
TH1 *input_hist0 =
dynamic_cast<TH1 *
>(folder->
FindObject(observablename.c_str()));
file->GetObject(samplelist[1].c_str(), folder);
TH1 *input_hist1 =
dynamic_cast<TH1 *
>(folder->
FindObject(observablename.c_str()));
file->GetObject(samplelist[2].c_str(), folder);
TH1 *input_hist2 =
dynamic_cast<TH1 *
>(folder->
FindObject(observablename.c_str()));
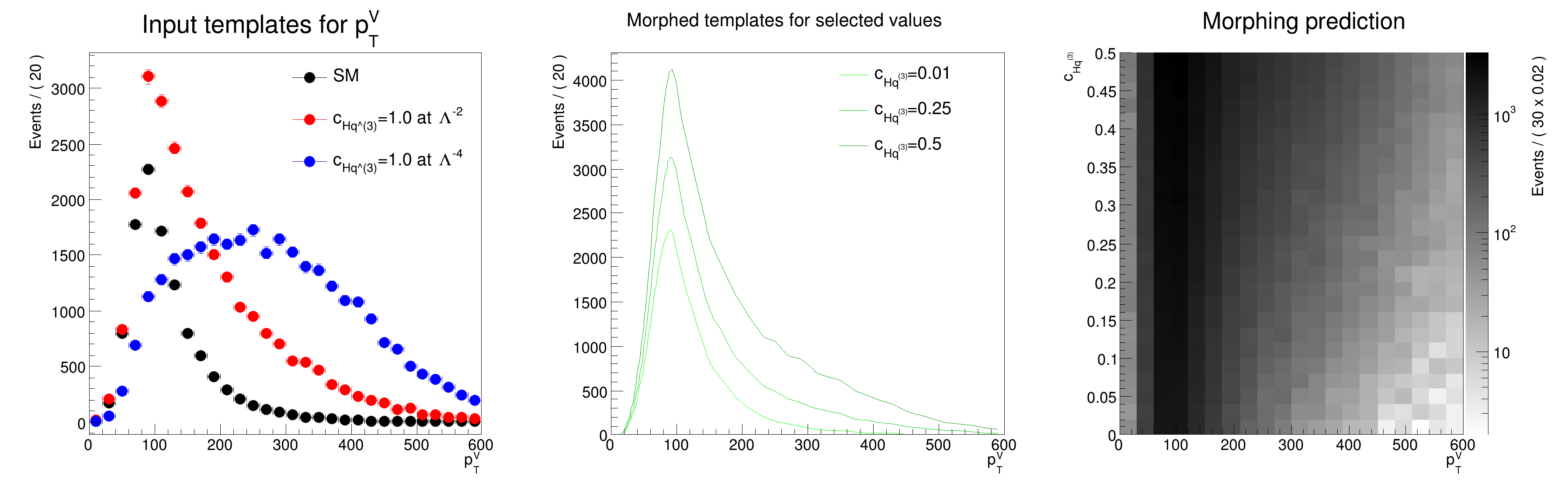
auto frame0 = obsvar.frame(Title("Input templates for p_{T}^{V}"));
input_dh0.plotOn(frame0, Name(samplelist[0].c_str()), LineColor(
kBlack), MarkerColor(
kBlack), MarkerSize(1));
input_dh1.plotOn(frame0, Name(samplelist[1].c_str()), LineColor(
kRed), MarkerColor(
kRed), MarkerSize(1));
input_dh2.plotOn(frame0, Name(samplelist[2].c_str()), LineColor(
kBlue), MarkerColor(
kBlue), MarkerSize(1));
auto frame1 = obsvar.frame(Title("Morphed templates for selected values"));
morph_datahist_0p01.plotOn(frame1, Name(
"morph_dh_cHq3=0.01"), DrawOption(
"C"), LineColor(
kGreen),
morph_datahist_0p25.plotOn(frame1, Name(
"morph_dh_cHq3=0.25"), DrawOption(
"C"), LineColor(
kGreen + 1),
morph_datahist_0p5.plotOn(frame1, Name(
"morph_dh_cHq3=0.5"), DrawOption(
"C"), LineColor(
kGreen + 2),
TH1 *hh_data = data->
createHistogram(
"pTV vs cHq3", obsvar, Binning(20), YVar(cHq3, Binning(50)));
hh_data->
SetTitle(
"Morphing prediction");
gPad->SetLeftMargin(0.15);
gPad->SetRightMargin(0.05);
frame0->Draw();
leg1->
AddEntry(frame0->findObject(
"SM_NPsq0"),
"SM",
"LP");
leg1->
AddEntry(frame0->findObject(
"cHq3_NPsq1"),
"c_{Hq^(3)}=1.0 at #Lambda^{-2}",
"LP");
leg1->
AddEntry(frame0->findObject(
"cHq3_NPsq2"),
"c_{Hq^(3)}=1.0 at #Lambda^{-4}",
"LP");
gPad->SetLeftMargin(0.15);
gPad->SetRightMargin(0.05);
frame1->Draw();
leg2->
AddEntry(frame1->findObject(
"morph_dh_cHq3=0.01"),
"c_{Hq^{(3)}}=0.01",
"L");
leg2->
AddEntry(frame1->findObject(
"morph_dh_cHq3=0.25"),
"c_{Hq^{(3)}}=0.25",
"L");
leg2->
AddEntry(frame1->findObject(
"morph_dh_cHq3=0.5"),
"c_{Hq^{(3)}}=0.5",
"L");
gPad->SetLeftMargin(0.12);
gPad->SetRightMargin(0.18);
c1->SaveAs(
"rf711_lagrangianmorph.png");
}
R__EXTERN TStyle * gStyle
virtual Bool_t add(const RooAbsArg &var, Bool_t silent=kFALSE)
Add the specified argument to list.
RooArgList is a container object that can hold multiple RooAbsArg objects.
RooArgSet is a container object that can hold multiple RooAbsArg objects.
The RooDataHist is a container class to hold N-dimensional binned data.
RooDataSet is a container class to hold unbinned data.
TH2F * createHistogram(const RooAbsRealLValue &var1, const RooAbsRealLValue &var2, const char *cuts="", const char *name="hist") const
Create a TH2F histogram of the distribution of the specified variable using this dataset.
Class RooLagrangianMorphing is a implementation of the method of Effective Lagrangian Morphing,...
RooRealVar represents a variable that can be changed from the outside.
The RooWrapperPdf is a class that can be used to convert a function into a PDF.
virtual void SetTitleOffset(Float_t offset=1)
Set distance between the axis and the axis title.
virtual void SetFillColor(Color_t fcolor)
Set the fill area color.
virtual void SetLineColor(Color_t lcolor)
Set the line color.
virtual void SetTextSize(Float_t tsize=1)
Set the text size.
virtual void SetRangeUser(Double_t ufirst, Double_t ulast)
Set the viewing range for the axis from ufirst to ulast (in user coordinates, that is,...
static void InvertPalette()
Invert the current color palette.
A ROOT file is a suite of consecutive data records (TKey instances) with a well defined format.
A TFolder object is a collection of objects and folders.
virtual TObject * FindObject(const char *name) const
Search object identified by name in the tree of folders inside this folder.
TH1 is the base class of all histogram classes in ROOT.
virtual void SetDirectory(TDirectory *dir)
By default, when a histogram is created, it is added to the list of histogram objects in the current ...
virtual void SetTitle(const char *title)
See GetStatOverflows for more information.
virtual void Draw(Option_t *option="")
Draw this histogram with options.
This class displays a legend box (TPaveText) containing several legend entries.
TLegendEntry * AddEntry(const TObject *obj, const char *label="", Option_t *option="lpf")
Add a new entry to this legend.
virtual void Draw(Option_t *option="")
Draw this legend with its current attributes.
virtual void SetTitle(const char *title="")
Set the title of the TNamed.
Mother of all ROOT objects.
void SetOptStat(Int_t stat=1)
The type of information printed in the histogram statistics box can be selected via the parameter mod...
void SetPalette(Int_t ncolors=kBird, Int_t *colors=0, Float_t alpha=1.)
See TColor::SetPalette.
void SetNumberContours(Int_t number=20)
Set the default number of contour levels when drawing 2-d plots.
The namespace RooFit contains mostly switches that change the behaviour of functions of PDFs (or othe...
std::string observableName
std::vector< std::string > folderNames

 Morphing effective field theory distributions with RooLagrangianMorphFunc A morphing function as a function of one coefficient is setup and can be used to obtain the distribution for any value of the coefficient.
Morphing effective field theory distributions with RooLagrangianMorphFunc A morphing function as a function of one coefficient is setup and can be used to obtain the distribution for any value of the coefficient.