␛[1mRooFit v3.60 -- Developed by Wouter Verkerke and David Kirkby␛[0m
Copyright (C) 2000-2013 NIKHEF, University of California & Stanford University
All rights reserved, please read http://roofit.sourceforge.net/license.txt
[#0] WARNING:InputArguments -- The parameter 'sigG' with range [-10, 10] of the RooGaussian 'g' exceeds the safe range of (0, inf). Advise to limit its range.
[#1] INFO:Minimization -- p.d.f. provides expected number of events, including extended term in likelihood.
[#1] INFO:Minimization -- RooAbsMinimizerFcn::setOptimizeConst: activating const optimization
[#1] INFO:Minimization -- The following expressions have been identified as constant and will be precalculated and cached: (p_h_sig,p_h_bkg)
**********
** 1 **SET PRINT 0
**********
**********
** 2 **SET NOGRAD
**********
PARAMETER DEFINITIONS:
NO. NAME VALUE STEP SIZE LIMITS
1 Abkg 1.00000e+00 4.95000e-01 1.00000e-02 5.00000e+03
2 Asig 1.00000e+00 4.95000e-01 1.00000e-02 5.00000e+03
**********
** 3 **SET ERR 0.5
**********
**********
** 4 **SET PRINT 0
**********
**********
** 5 **SET STR 1
**********
**********
** 6 **MIGRAD 1000 1
**********
MIGRAD MINIMIZATION HAS CONVERGED.
MIGRAD WILL VERIFY CONVERGENCE AND ERROR MATRIX.
FCN=-2388.31 FROM MIGRAD STATUS=CONVERGED 59 CALLS 60 TOTAL
EDM=6.50963e-06 STRATEGY= 1 ERROR MATRIX ACCURATE
EXT PARAMETER STEP FIRST
NO. NAME VALUE ERROR SIZE DERIVATIVE
1 Abkg 6.14547e-02 2.11669e-03 4.23105e-06 -2.89246e+00
2 Asig 8.33778e-01 1.89800e-01 9.38840e-05 -8.95027e-01
ERR DEF= 0.5
**********
** 7 **SET ERR 0.5
**********
**********
** 8 **SET PRINT 0
**********
**********
** 9 **HESSE 1000
**********
FCN=-2388.31 FROM HESSE STATUS=OK 10 CALLS 70 TOTAL
EDM=6.51699e-06 STRATEGY= 1 ERROR MATRIX ACCURATE
EXT PARAMETER INTERNAL INTERNAL
NO. NAME VALUE ERROR STEP SIZE VALUE
1 Abkg 6.14547e-02 2.11685e-03 8.46210e-07 -1.56438e+00
2 Asig 8.33778e-01 1.89814e-01 1.87768e-05 -1.54512e+00
ERR DEF= 0.5
[#1] INFO:Minimization -- RooAbsMinimizerFcn::setOptimizeConst: deactivating const optimization
[#1] INFO:Minimization -- p.d.f. provides expected number of events, including extended term in likelihood.
[#1] INFO:Minimization -- Including the following constraint terms in minimization: (hc_sig,hc_bkg)
[#1] INFO:Minimization -- The global observables are not defined , normalize constraints with respect to the parameters (Abkg,Asig,p_ph_bkg_gamma_bin_0,p_ph_bkg_gamma_bin_1,p_ph_bkg_gamma_bin_10,p_ph_bkg_gamma_bin_11,p_ph_bkg_gamma_bin_12,p_ph_bkg_gamma_bin_13,p_ph_bkg_gamma_bin_14,p_ph_bkg_gamma_bin_15,p_ph_bkg_gamma_bin_16,p_ph_bkg_gamma_bin_17,p_ph_bkg_gamma_bin_18,p_ph_bkg_gamma_bin_19,p_ph_bkg_gamma_bin_2,p_ph_bkg_gamma_bin_20,p_ph_bkg_gamma_bin_21,p_ph_bkg_gamma_bin_22,p_ph_bkg_gamma_bin_23,p_ph_bkg_gamma_bin_24,p_ph_bkg_gamma_bin_3,p_ph_bkg_gamma_bin_4,p_ph_bkg_gamma_bin_5,p_ph_bkg_gamma_bin_6,p_ph_bkg_gamma_bin_7,p_ph_bkg_gamma_bin_8,p_ph_bkg_gamma_bin_9,p_ph_sig_gamma_bin_0,p_ph_sig_gamma_bin_1,p_ph_sig_gamma_bin_10,p_ph_sig_gamma_bin_11,p_ph_sig_gamma_bin_12,p_ph_sig_gamma_bin_13,p_ph_sig_gamma_bin_14,p_ph_sig_gamma_bin_15,p_ph_sig_gamma_bin_16,p_ph_sig_gamma_bin_17,p_ph_sig_gamma_bin_18,p_ph_sig_gamma_bin_19,p_ph_sig_gamma_bin_2,p_ph_sig_gamma_bin_20,p_ph_sig_gamma_bin_21,p_ph_sig_gamma_bin_22,p_ph_sig_gamma_bin_23,p_ph_sig_gamma_bin_24,p_ph_sig_gamma_bin_3,p_ph_sig_gamma_bin_4,p_ph_sig_gamma_bin_5,p_ph_sig_gamma_bin_6,p_ph_sig_gamma_bin_7,p_ph_sig_gamma_bin_8,p_ph_sig_gamma_bin_9)
[#1] INFO:Fitting -- RooAddition::defaultErrorLevel(nll_model1_sumData_with_constr) Summation contains a RooNLLVar, using its error level
[#1] INFO:Minimization -- RooAbsMinimizerFcn::setOptimizeConst: activating const optimization
[#1] INFO:Minimization -- The following expressions will be evaluated in cache-and-track mode: (p_ph_sig,p_ph_bkg)
**********
** 1 **SET PRINT 0
**********
**********
** 2 **SET NOGRAD
**********
PARAMETER DEFINITIONS:
NO. NAME VALUE STEP SIZE LIMITS
1 Abkg 1.00000e+00 4.95000e-01 1.00000e-02 5.00000e+03
2 Asig 1.00000e+00 4.95000e-01 1.00000e-02 5.00000e+03
3 p_ph_bkg_gamma_bin_0 1.00000e+00 4.96292e-02 0.00000e+00 1.00000e+03
4 p_ph_bkg_gamma_bin_1 1.00000e+00 4.96292e-02 0.00000e+00 1.00000e+03
5 p_ph_bkg_gamma_bin_10 1.00000e+00 4.81683e-02 0.00000e+00 1.00000e+03
6 p_ph_bkg_gamma_bin_11 1.00000e+00 5.17088e-02 0.00000e+00 1.00000e+03
7 p_ph_bkg_gamma_bin_12 1.00000e+00 5.11645e-02 0.00000e+00 1.00000e+03
8 p_ph_bkg_gamma_bin_13 1.00000e+00 5.01255e-02 0.00000e+00 1.00000e+03
9 p_ph_bkg_gamma_bin_14 1.00000e+00 5.13665e-02 0.00000e+00 1.00000e+03
10 p_ph_bkg_gamma_bin_15 1.00000e+00 4.93865e-02 0.00000e+00 1.00000e+03
11 p_ph_bkg_gamma_bin_16 1.00000e+00 5.07020e-02 0.00000e+00 1.00000e+03
12 p_ph_bkg_gamma_bin_17 1.00000e+00 4.86792e-02 0.00000e+00 1.00000e+03
13 p_ph_bkg_gamma_bin_18 1.00000e+00 5.27780e-02 0.00000e+00 1.00000e+03
14 p_ph_bkg_gamma_bin_19 1.00000e+00 4.85643e-02 0.00000e+00 1.00000e+03
15 p_ph_bkg_gamma_bin_2 1.00000e+00 4.90881e-02 0.00000e+00 1.00000e+03
16 p_ph_bkg_gamma_bin_20 1.00000e+00 4.95074e-02 0.00000e+00 1.00000e+03
17 p_ph_bkg_gamma_bin_21 1.00000e+00 4.96904e-02 0.00000e+00 1.00000e+03
18 p_ph_bkg_gamma_bin_22 1.00000e+00 5.10310e-02 0.00000e+00 1.00000e+03
19 p_ph_bkg_gamma_bin_23 1.00000e+00 4.86792e-02 0.00000e+00 1.00000e+03
20 p_ph_bkg_gamma_bin_24 1.00000e+00 4.95074e-02 0.00000e+00 1.00000e+03
21 p_ph_bkg_gamma_bin_3 1.00000e+00 5.17088e-02 0.00000e+00 1.00000e+03
22 p_ph_bkg_gamma_bin_4 1.00000e+00 5.08329e-02 0.00000e+00 1.00000e+03
23 p_ph_bkg_gamma_bin_5 1.00000e+00 5.06370e-02 0.00000e+00 1.00000e+03
24 p_ph_bkg_gamma_bin_6 1.00000e+00 5.17088e-02 0.00000e+00 1.00000e+03
25 p_ph_bkg_gamma_bin_7 1.00000e+00 4.81683e-02 0.00000e+00 1.00000e+03
26 p_ph_bkg_gamma_bin_8 1.00000e+00 4.89116e-02 0.00000e+00 1.00000e+03
27 p_ph_bkg_gamma_bin_9 1.00000e+00 4.97519e-02 0.00000e+00 1.00000e+03
28 p_ph_sig_gamma_bin_11 1.00000e+00 3.53553e-01 0.00000e+00 1.00000e+03
29 p_ph_sig_gamma_bin_12 1.00000e+00 2.35702e-01 0.00000e+00 1.00000e+03
30 p_ph_sig_gamma_bin_13 1.00000e+00 2.42536e-01 0.00000e+00 1.00000e+03
31 p_ph_sig_gamma_bin_14 1.00000e+00 4.08248e-01 0.00000e+00 1.00000e+03
32 p_ph_sig_gamma_bin_15 1.00000e+00 1.00000e+00 0.00000e+00 1.00000e+03
MINUIT WARNING IN PARAMETR
============== VARIABLE32 IS AT ITS LOWER ALLOWED LIMIT.
**********
** 3 **SET ERR 0.5
**********
**********
** 4 **SET PRINT 0
**********
**********
** 5 **SET STR 1
**********
**********
** 6 **MIGRAD 1.6e+04 1
**********
MIGRAD MINIMIZATION HAS CONVERGED.
FCN=-2282.23 FROM MIGRAD STATUS=CONVERGED 678 CALLS 679 TOTAL
EDM=3.22904e-05 STRATEGY= 1 ERROR MATRIX UNCERTAINTY 4.0 per cent
EXT PARAMETER STEP FIRST
NO. NAME VALUE ERROR SIZE DERIVATIVE
1 Abkg 6.13800e-02 2.20007e-03 1.23597e-06 2.83314e+01
2 Asig 8.50507e-01 2.30325e-01 6.38421e-05 5.87286e-01
3 p_ph_bkg_gamma_bin_0 9.98047e-01 3.71725e-02 -7.16183e-06 5.44794e-02
4 p_ph_bkg_gamma_bin_1 1.00029e+00 3.72063e-02 -6.99607e-06 6.61256e-02
5 p_ph_bkg_gamma_bin_10 9.80291e-01 3.58648e-02 -8.43676e-06 -4.39623e-02
6 p_ph_bkg_gamma_bin_11 1.00612e+00 3.95761e-02 -1.30946e-05 -2.40671e+00
7 p_ph_bkg_gamma_bin_12 1.00453e+00 4.49136e-02 2.72725e-05 -2.74507e+00
8 p_ph_bkg_gamma_bin_13 9.91157e-01 4.00819e-02 1.95392e-05 -5.92968e-01
9 p_ph_bkg_gamma_bin_14 9.95972e-01 3.84117e-02 -7.35477e-06 -9.64110e-01
10 p_ph_bkg_gamma_bin_15 1.00900e+00 3.73278e-02 -1.24049e-05 2.95046e-01
11 p_ph_bkg_gamma_bin_16 1.02997e+00 3.84551e-02 -4.71081e-06 2.00609e-01
12 p_ph_bkg_gamma_bin_17 1.00768e+00 3.66321e-02 -6.44293e-06 1.07790e-01
13 p_ph_bkg_gamma_bin_18 1.01965e+00 3.97945e-02 -5.52428e-06 1.43183e-01
14 p_ph_bkg_gamma_bin_19 9.74997e-01 3.60743e-02 -8.80379e-06 -7.43750e-02
15 p_ph_bkg_gamma_bin_2 9.96150e-01 3.67541e-02 -7.30122e-06 4.55320e-02
16 p_ph_bkg_gamma_bin_20 9.99851e-01 3.71118e-02 -7.02865e-06 6.41653e-02
17 p_ph_bkg_gamma_bin_21 9.98263e-01 3.72199e-02 -7.14591e-06 5.54687e-02
18 p_ph_bkg_gamma_bin_22 9.98316e-01 3.81884e-02 -7.14203e-06 5.28519e-02
19 p_ph_bkg_gamma_bin_23 9.83937e-01 3.62827e-02 -8.18043e-06 -2.20353e-02
20 p_ph_bkg_gamma_bin_24 1.02664e+00 3.75311e-02 -4.97535e-06 1.95313e-01
21 p_ph_bkg_gamma_bin_3 1.00553e+00 3.87905e-02 -6.60476e-06 8.55876e-02
22 p_ph_bkg_gamma_bin_4 9.90574e-01 3.79282e-02 -7.70648e-06 1.42148e-02
23 p_ph_bkg_gamma_bin_5 9.92294e-01 3.78128e-02 -7.58214e-06 2.31734e-02
24 p_ph_bkg_gamma_bin_6 1.01041e+00 3.88677e-02 -6.23632e-06 1.08003e-01
25 p_ph_bkg_gamma_bin_7 9.97199e-01 3.61069e-02 -7.22424e-06 5.31287e-02
26 p_ph_bkg_gamma_bin_8 9.80281e-01 3.63974e-02 -8.43747e-06 -4.26945e-02
27 p_ph_bkg_gamma_bin_9 1.00975e+00 3.74398e-02 -6.28583e-06 1.13462e-01
28 p_ph_sig_gamma_bin_11 1.09459e+00 2.95086e-01 -2.14193e-04 -1.90672e-01
29 p_ph_sig_gamma_bin_12 1.07095e+00 2.09961e-01 -8.87826e-05 3.71203e-01
30 p_ph_sig_gamma_bin_13 8.91495e-01 1.94710e-01 -4.27837e-05 6.41259e-01
31 p_ph_sig_gamma_bin_14 9.47683e-01 3.09459e-01 -1.41412e-04 -5.27753e-02
32 p_ph_sig_gamma_bin_15 1.14095e+00 8.19493e-01 1.63221e-04 8.16586e-03
ERR DEF= 0.5
**********
** 7 **SET ERR 0.5
**********
**********
** 8 **SET PRINT 0
**********
**********
** 9 **HESSE 1.6e+04
**********
FCN=-2282.23 FROM HESSE STATUS=OK 639 CALLS 1318 TOTAL
EDM=7.72255e-05 STRATEGY= 1 ERROR MATRIX ACCURATE
EXT PARAMETER INTERNAL INTERNAL
NO. NAME VALUE ERROR STEP SIZE VALUE
1 Abkg 6.13800e-02 2.22709e-03 4.12889e-06 -1.56439e+00
2 Asig 8.50507e-01 2.35778e-01 9.17056e-05 -1.54486e+00
3 p_ph_bkg_gamma_bin_0 9.98047e-01 4.74228e-02 4.94370e-05 -1.50760e+00
4 p_ph_bkg_gamma_bin_1 1.00029e+00 4.74763e-02 4.94370e-05 -1.50753e+00
5 p_ph_bkg_gamma_bin_10 9.80291e-01 4.56200e-02 4.79818e-05 -1.50817e+00
6 p_ph_bkg_gamma_bin_11 1.00612e+00 5.08524e-02 5.21176e-05 -1.50735e+00
7 p_ph_bkg_gamma_bin_12 1.00453e+00 5.09701e-02 5.19588e-05 -1.50740e+00
8 p_ph_bkg_gamma_bin_13 9.91157e-01 4.89292e-02 5.06394e-05 -1.50782e+00
9 p_ph_bkg_gamma_bin_14 9.95972e-01 4.97163e-02 5.15499e-05 -1.50767e+00
10 p_ph_bkg_gamma_bin_15 1.00900e+00 4.76423e-02 4.92850e-05 -1.50726e+00
11 p_ph_bkg_gamma_bin_16 1.02997e+00 4.92155e-02 5.05056e-05 -1.50660e+00
12 p_ph_bkg_gamma_bin_17 1.00768e+00 4.67445e-02 4.84907e-05 -1.50730e+00
13 p_ph_bkg_gamma_bin_18 1.01965e+00 5.09631e-02 5.25736e-05 -1.50692e+00
14 p_ph_bkg_gamma_bin_19 9.74997e-01 4.58683e-02 4.83762e-05 -1.50834e+00
15 p_ph_bkg_gamma_bin_2 9.96150e-01 4.68633e-02 4.88980e-05 -1.50766e+00
16 p_ph_bkg_gamma_bin_20 9.99851e-01 4.73499e-02 4.93157e-05 -1.50754e+00
17 p_ph_bkg_gamma_bin_21 9.98263e-01 4.74862e-02 4.94980e-05 -1.50760e+00
18 p_ph_bkg_gamma_bin_22 9.98316e-01 4.87627e-02 5.08334e-05 -1.50759e+00
19 p_ph_bkg_gamma_bin_23 9.83937e-01 4.61877e-02 4.84907e-05 -1.50805e+00
20 p_ph_bkg_gamma_bin_24 1.02664e+00 4.79832e-02 4.93156e-05 -1.50670e+00
21 p_ph_bkg_gamma_bin_3 1.00553e+00 4.95866e-02 5.15085e-05 -1.50737e+00
22 p_ph_bkg_gamma_bin_4 9.90574e-01 4.83846e-02 5.06360e-05 -1.50784e+00
23 p_ph_bkg_gamma_bin_5 9.92294e-01 4.82410e-02 5.04409e-05 -1.50778e+00
24 p_ph_bkg_gamma_bin_6 1.01041e+00 4.97071e-02 5.15085e-05 -1.50721e+00
25 p_ph_bkg_gamma_bin_7 9.97199e-01 4.60138e-02 4.79818e-05 -1.50763e+00
26 p_ph_bkg_gamma_bin_8 9.80281e-01 4.63204e-02 4.87222e-05 -1.50817e+00
27 p_ph_bkg_gamma_bin_9 1.00975e+00 4.78188e-02 4.95592e-05 -1.50723e+00
28 p_ph_sig_gamma_bin_11 1.09459e+00 3.37438e-01 3.28479e-04 -1.50462e+00
29 p_ph_sig_gamma_bin_12 1.07095e+00 2.21326e-01 1.99896e-04 -1.50533e+00
30 p_ph_sig_gamma_bin_13 8.91495e-01 2.03106e-01 2.03458e-04 -1.51107e+00
31 p_ph_sig_gamma_bin_14 9.47683e-01 3.57655e-01 3.77115e-04 -1.50922e+00
32 p_ph_sig_gamma_bin_15 1.14095e+00 1.11196e+00 1.08088e-03 -1.50323e+00
ERR DEF= 0.5
[#1] INFO:Minimization -- RooAbsMinimizerFcn::setOptimizeConst: deactivating const optimization
[#1] INFO:Minimization -- p.d.f. provides expected number of events, including extended term in likelihood.
[#1] INFO:Minimization -- Including the following constraint terms in minimization: (hc_sigbkg)
[#1] INFO:Minimization -- The global observables are not defined , normalize constraints with respect to the parameters (Abkg,Asig,p_ph_sig2_gamma_bin_0,p_ph_sig2_gamma_bin_1,p_ph_sig2_gamma_bin_10,p_ph_sig2_gamma_bin_11,p_ph_sig2_gamma_bin_12,p_ph_sig2_gamma_bin_13,p_ph_sig2_gamma_bin_14,p_ph_sig2_gamma_bin_15,p_ph_sig2_gamma_bin_16,p_ph_sig2_gamma_bin_17,p_ph_sig2_gamma_bin_18,p_ph_sig2_gamma_bin_19,p_ph_sig2_gamma_bin_2,p_ph_sig2_gamma_bin_20,p_ph_sig2_gamma_bin_21,p_ph_sig2_gamma_bin_22,p_ph_sig2_gamma_bin_23,p_ph_sig2_gamma_bin_24,p_ph_sig2_gamma_bin_3,p_ph_sig2_gamma_bin_4,p_ph_sig2_gamma_bin_5,p_ph_sig2_gamma_bin_6,p_ph_sig2_gamma_bin_7,p_ph_sig2_gamma_bin_8,p_ph_sig2_gamma_bin_9)
[#1] INFO:Fitting -- RooAddition::defaultErrorLevel(nll_model2_sumData_with_constr) Summation contains a RooNLLVar, using its error level
[#1] INFO:Minimization -- RooAbsMinimizerFcn::setOptimizeConst: activating const optimization
[#1] INFO:Minimization -- The following expressions will be evaluated in cache-and-track mode: (p_ph_sig2,p_ph_bkg2)
**********
** 1 **SET PRINT 0
**********
**********
** 2 **SET NOGRAD
**********
PARAMETER DEFINITIONS:
NO. NAME VALUE STEP SIZE LIMITS
1 Abkg 1.00000e+00 4.95000e-01 1.00000e-02 5.00000e+03
2 Asig 1.00000e+00 4.95000e-01 1.00000e-02 5.00000e+03
3 p_ph_sig2_gamma_bin_0 1.00000e+00 inf 0.00000e+00 1.00000e+03
MINUIT WARNING IN PARAMETR
============== VARIABLE3 BROUGHT BACK INSIDE LIMITS.
MINUIT WARNING IN PARAMETR
============== VARIABLE3 BROUGHT BACK INSIDE LIMITS.
4 p_ph_sig2_gamma_bin_1 1.00000e+00 inf 0.00000e+00 1.00000e+03
MINUIT WARNING IN PARAMETR
============== VARIABLE4 BROUGHT BACK INSIDE LIMITS.
MINUIT WARNING IN PARAMETR
============== VARIABLE4 BROUGHT BACK INSIDE LIMITS.
5 p_ph_sig2_gamma_bin_10 1.00000e+00 inf 0.00000e+00 1.00000e+03
MINUIT WARNING IN PARAMETR
============== VARIABLE5 BROUGHT BACK INSIDE LIMITS.
MINUIT WARNING IN PARAMETR
============== VARIABLE5 BROUGHT BACK INSIDE LIMITS.
6 p_ph_sig2_gamma_bin_11 1.00000e+00 3.53553e-01 0.00000e+00 1.00000e+03
7 p_ph_sig2_gamma_bin_12 1.00000e+00 2.35702e-01 0.00000e+00 1.00000e+03
8 p_ph_sig2_gamma_bin_13 1.00000e+00 2.42536e-01 0.00000e+00 1.00000e+03
9 p_ph_sig2_gamma_bin_14 1.00000e+00 4.08248e-01 0.00000e+00 1.00000e+03
10 p_ph_sig2_gamma_bin_15 1.00000e+00 1.00000e+00 0.00000e+00 1.00000e+03
MINUIT WARNING IN PARAMETR
============== VARIABLE10 IS AT ITS LOWER ALLOWED LIMIT.
11 p_ph_sig2_gamma_bin_16 1.00000e+00 inf 0.00000e+00 1.00000e+03
MINUIT WARNING IN PARAMETR
============== VARIABLE11 BROUGHT BACK INSIDE LIMITS.
MINUIT WARNING IN PARAMETR
============== VARIABLE11 BROUGHT BACK INSIDE LIMITS.
12 p_ph_sig2_gamma_bin_17 1.00000e+00 inf 0.00000e+00 1.00000e+03
MINUIT WARNING IN PARAMETR
============== VARIABLE12 BROUGHT BACK INSIDE LIMITS.
MINUIT WARNING IN PARAMETR
============== VARIABLE12 BROUGHT BACK INSIDE LIMITS.
13 p_ph_sig2_gamma_bin_18 1.00000e+00 inf 0.00000e+00 1.00000e+03
MINUIT WARNING IN PARAMETR
============== VARIABLE13 BROUGHT BACK INSIDE LIMITS.
MINUIT WARNING IN PARAMETR
============== VARIABLE13 BROUGHT BACK INSIDE LIMITS.
14 p_ph_sig2_gamma_bin_19 1.00000e+00 inf 0.00000e+00 1.00000e+03
MINUIT WARNING IN PARAMETR
============== VARIABLE14 BROUGHT BACK INSIDE LIMITS.
MINUIT WARNING IN PARAMETR
============== VARIABLE14 BROUGHT BACK INSIDE LIMITS.
15 p_ph_sig2_gamma_bin_2 1.00000e+00 inf 0.00000e+00 1.00000e+03
MINUIT WARNING IN PARAMETR
============== VARIABLE15 BROUGHT BACK INSIDE LIMITS.
MINUIT WARNING IN PARAMETR
============== VARIABLE15 BROUGHT BACK INSIDE LIMITS.
16 p_ph_sig2_gamma_bin_20 1.00000e+00 inf 0.00000e+00 1.00000e+03
MINUIT WARNING IN PARAMETR
============== VARIABLE16 BROUGHT BACK INSIDE LIMITS.
MINUIT WARNING IN PARAMETR
============== VARIABLE16 BROUGHT BACK INSIDE LIMITS.
17 p_ph_sig2_gamma_bin_21 1.00000e+00 inf 0.00000e+00 1.00000e+03
MINUIT WARNING IN PARAMETR
============== VARIABLE17 BROUGHT BACK INSIDE LIMITS.
MINUIT WARNING IN PARAMETR
============== VARIABLE17 BROUGHT BACK INSIDE LIMITS.
18 p_ph_sig2_gamma_bin_22 1.00000e+00 inf 0.00000e+00 1.00000e+03
MINUIT WARNING IN PARAMETR
============== VARIABLE18 BROUGHT BACK INSIDE LIMITS.
MINUIT WARNING IN PARAMETR
============== VARIABLE18 BROUGHT BACK INSIDE LIMITS.
19 p_ph_sig2_gamma_bin_23 1.00000e+00 inf 0.00000e+00 1.00000e+03
MINUIT WARNING IN PARAMETR
============== VARIABLE19 BROUGHT BACK INSIDE LIMITS.
MINUIT WARNING IN PARAMETR
============== VARIABLE19 BROUGHT BACK INSIDE LIMITS.
20 p_ph_sig2_gamma_bin_24 1.00000e+00 inf 0.00000e+00 1.00000e+03
MINUIT WARNING IN PARAMETR
============== VARIABLE20 BROUGHT BACK INSIDE LIMITS.
MINUIT WARNING IN PARAMETR
============== VARIABLE20 BROUGHT BACK INSIDE LIMITS.
21 p_ph_sig2_gamma_bin_3 1.00000e+00 inf 0.00000e+00 1.00000e+03
MINUIT WARNING IN PARAMETR
============== VARIABLE21 BROUGHT BACK INSIDE LIMITS.
MINUIT WARNING IN PARAMETR
============== VARIABLE21 BROUGHT BACK INSIDE LIMITS.
22 p_ph_sig2_gamma_bin_4 1.00000e+00 inf 0.00000e+00 1.00000e+03
MINUIT WARNING IN PARAMETR
============== VARIABLE22 BROUGHT BACK INSIDE LIMITS.
MINUIT WARNING IN PARAMETR
============== VARIABLE22 BROUGHT BACK INSIDE LIMITS.
23 p_ph_sig2_gamma_bin_5 1.00000e+00 inf 0.00000e+00 1.00000e+03
MINUIT WARNING IN PARAMETR
============== VARIABLE23 BROUGHT BACK INSIDE LIMITS.
MINUIT WARNING IN PARAMETR
============== VARIABLE23 BROUGHT BACK INSIDE LIMITS.
24 p_ph_sig2_gamma_bin_6 1.00000e+00 inf 0.00000e+00 1.00000e+03
MINUIT WARNING IN PARAMETR
============== VARIABLE24 BROUGHT BACK INSIDE LIMITS.
MINUIT WARNING IN PARAMETR
============== VARIABLE24 BROUGHT BACK INSIDE LIMITS.
25 p_ph_sig2_gamma_bin_7 1.00000e+00 inf 0.00000e+00 1.00000e+03
MINUIT WARNING IN PARAMETR
============== VARIABLE25 BROUGHT BACK INSIDE LIMITS.
MINUIT WARNING IN PARAMETR
============== VARIABLE25 BROUGHT BACK INSIDE LIMITS.
26 p_ph_sig2_gamma_bin_8 1.00000e+00 inf 0.00000e+00 1.00000e+03
MINUIT WARNING IN PARAMETR
============== VARIABLE26 BROUGHT BACK INSIDE LIMITS.
MINUIT WARNING IN PARAMETR
============== VARIABLE26 BROUGHT BACK INSIDE LIMITS.
27 p_ph_sig2_gamma_bin_9 1.00000e+00 inf 0.00000e+00 1.00000e+03
MINUIT WARNING IN PARAMETR
============== VARIABLE27 BROUGHT BACK INSIDE LIMITS.
MINUIT WARNING IN PARAMETR
============== VARIABLE27 BROUGHT BACK INSIDE LIMITS.
**********
** 3 **SET ERR 0.5
**********
**********
** 4 **SET PRINT 0
**********
**********
** 5 **SET STR 1
**********
**********
** 6 **MIGRAD 1.35e+04 1
**********
MIGRAD MINIMIZATION HAS CONVERGED.
MIGRAD WILL VERIFY CONVERGENCE AND ERROR MATRIX.
FCN=-2291.45 FROM MIGRAD STATUS=CONVERGED 1074 CALLS 1075 TOTAL
EDM=5.33608e-05 STRATEGY= 1 ERROR MATRIX ACCURATE
EXT PARAMETER STEP FIRST
NO. NAME VALUE ERROR SIZE DERIVATIVE
1 Abkg 6.14396e-02 2.21881e-03 4.13559e-06 -4.41296e+01
2 Asig 8.34408e-01 2.03008e-01 9.30538e-05 -8.65120e-01
3 p_ph_sig2_gamma_bin_0 9.97977e-01 4.74184e-02 4.95146e-05 2.82438e-01
4 p_ph_sig2_gamma_bin_1 1.00022e+00 4.74717e-02 4.95144e-05 2.30502e-01
5 p_ph_sig2_gamma_bin_10 9.80256e-01 4.56170e-02 4.80590e-05 7.14152e-01
6 p_ph_sig2_gamma_bin_11 1.01066e+00 4.88753e-02 5.04866e-05 8.12095e-01
7 p_ph_sig2_gamma_bin_12 1.01175e+00 4.83913e-02 4.87304e-05 1.45863e-01
8 p_ph_sig2_gamma_bin_13 9.83606e-01 4.66986e-02 4.79414e-05 1.21197e-01
9 p_ph_sig2_gamma_bin_14 9.94795e-01 4.83050e-02 5.04315e-05 8.78700e-01
10 p_ph_sig2_gamma_bin_15 1.00964e+00 4.73305e-02 4.91474e-05 1.06952e-01
11 p_ph_sig2_gamma_bin_16 1.02983e+00 4.92085e-02 5.05815e-05 -4.88517e-01
12 p_ph_sig2_gamma_bin_17 1.00759e+00 4.67394e-02 4.85659e-05 5.74385e-02
13 p_ph_sig2_gamma_bin_18 1.01953e+00 5.09568e-02 5.26537e-05 -2.13148e-01
14 p_ph_sig2_gamma_bin_19 9.74972e-01 4.58656e-02 4.84546e-05 8.16686e-01
15 p_ph_sig2_gamma_bin_2 9.96083e-01 4.68591e-02 4.89750e-05 3.33076e-01
16 p_ph_sig2_gamma_bin_20 9.99777e-01 4.73454e-02 4.93929e-05 2.41903e-01
17 p_ph_sig2_gamma_bin_21 9.98192e-01 4.74818e-02 4.95757e-05 2.76785e-01
18 p_ph_sig2_gamma_bin_22 9.98245e-01 4.87583e-02 5.09132e-05 2.61429e-01
19 p_ph_sig2_gamma_bin_23 9.83895e-01 4.61844e-02 4.85684e-05 6.19089e-01
20 p_ph_sig2_gamma_bin_24 1.02651e+00 4.79766e-02 4.93901e-05 -4.24164e-01
21 p_ph_sig2_gamma_bin_3 1.00545e+00 4.95815e-02 5.15886e-05 9.86817e-02
22 p_ph_sig2_gamma_bin_4 9.90519e-01 4.83807e-02 5.07164e-05 4.30253e-01
23 p_ph_sig2_gamma_bin_5 9.92236e-01 4.82370e-02 5.05207e-05 3.96794e-01
24 p_ph_sig2_gamma_bin_6 1.01031e+00 4.97016e-02 5.15880e-05 -9.57945e-03
25 p_ph_sig2_gamma_bin_7 9.97130e-01 4.60096e-02 4.80572e-05 3.20349e-01
26 p_ph_sig2_gamma_bin_8 9.80246e-01 4.63173e-02 4.88006e-05 6.92914e-01
27 p_ph_sig2_gamma_bin_9 1.00966e+00 4.78135e-02 4.96358e-05 5.22262e-03
ERR DEF= 0.5
**********
** 7 **SET ERR 0.5
**********
**********
** 8 **SET PRINT 0
**********
**********
** 9 **HESSE 1.35e+04
**********
FCN=-2291.45 FROM HESSE STATUS=OK 460 CALLS 1535 TOTAL
EDM=5.33547e-05 STRATEGY= 1 ERROR MATRIX ACCURATE
EXT PARAMETER INTERNAL INTERNAL
NO. NAME VALUE ERROR STEP SIZE VALUE
1 Abkg 6.14396e-02 2.21887e-03 8.27117e-07 -1.56438e+00
2 Asig 8.34408e-01 2.02972e-01 1.86108e-05 -1.54511e+00
3 p_ph_sig2_gamma_bin_0 9.97977e-01 4.74183e-02 9.90292e-06 -1.50760e+00
4 p_ph_sig2_gamma_bin_1 1.00022e+00 4.74716e-02 9.90288e-06 -1.50753e+00
5 p_ph_sig2_gamma_bin_10 9.80256e-01 4.56169e-02 9.61180e-06 -1.50817e+00
6 p_ph_sig2_gamma_bin_11 1.01066e+00 4.88739e-02 1.00973e-05 -1.50720e+00
7 p_ph_sig2_gamma_bin_12 1.01175e+00 4.83851e-02 9.74607e-06 -1.50717e+00
8 p_ph_sig2_gamma_bin_13 9.83606e-01 4.66936e-02 9.58827e-06 -1.50806e+00
9 p_ph_sig2_gamma_bin_14 9.94795e-01 4.83042e-02 1.00863e-05 -1.50771e+00
10 p_ph_sig2_gamma_bin_15 1.00964e+00 4.73303e-02 9.82949e-06 -1.50724e+00
11 p_ph_sig2_gamma_bin_16 1.02983e+00 4.92084e-02 1.01163e-05 -1.50660e+00
12 p_ph_sig2_gamma_bin_17 1.00759e+00 4.67393e-02 9.71318e-06 -1.50730e+00
13 p_ph_sig2_gamma_bin_18 1.01953e+00 5.09567e-02 1.05307e-05 -1.50693e+00
14 p_ph_sig2_gamma_bin_19 9.74972e-01 4.58655e-02 9.69093e-06 -1.50834e+00
15 p_ph_sig2_gamma_bin_2 9.96083e-01 4.68590e-02 9.79499e-06 -1.50766e+00
16 p_ph_sig2_gamma_bin_20 9.99777e-01 4.73452e-02 9.87858e-06 -1.50755e+00
17 p_ph_sig2_gamma_bin_21 9.98192e-01 4.74817e-02 9.91514e-06 -1.50760e+00
18 p_ph_sig2_gamma_bin_22 9.98245e-01 4.87581e-02 1.01826e-05 -1.50760e+00
19 p_ph_sig2_gamma_bin_23 9.83895e-01 4.61843e-02 9.71367e-06 -1.50805e+00
20 p_ph_sig2_gamma_bin_24 1.02651e+00 4.79765e-02 9.87802e-06 -1.50671e+00
21 p_ph_sig2_gamma_bin_3 1.00545e+00 4.95814e-02 1.03177e-05 -1.50737e+00
22 p_ph_sig2_gamma_bin_4 9.90519e-01 4.83806e-02 1.01433e-05 -1.50784e+00
23 p_ph_sig2_gamma_bin_5 9.92236e-01 4.82369e-02 1.01041e-05 -1.50779e+00
24 p_ph_sig2_gamma_bin_6 1.01031e+00 4.97015e-02 2.06352e-06 -1.50721e+00
25 p_ph_sig2_gamma_bin_7 9.97130e-01 4.60094e-02 9.61144e-06 -1.50763e+00
26 p_ph_sig2_gamma_bin_8 9.80246e-01 4.63172e-02 9.76012e-06 -1.50817e+00
27 p_ph_sig2_gamma_bin_9 1.00966e+00 4.78134e-02 1.98543e-06 -1.50724e+00
ERR DEF= 0.5
[#1] INFO:Minimization -- RooAbsMinimizerFcn::setOptimizeConst: deactivating const optimization
[#1] INFO:Plotting -- RooAbsPdf::plotOn(model0) directly selected PDF components: (p_h_sig)
[#1] INFO:Plotting -- RooAbsPdf::plotOn(model0) indirectly selected PDF components: ()
[#1] INFO:Plotting -- RooAbsPdf::plotOn(model0) directly selected PDF components: (p_h_bkg)
[#1] INFO:Plotting -- RooAbsPdf::plotOn(model0) indirectly selected PDF components: ()
[#1] INFO:Plotting -- RooPlot::updateFitRangeNorm: New event count of 50 will supercede previous event count of 1050 for normalization of PDF projections
[#1] INFO:Plotting -- RooAbsPdf::plotOn(model1) directly selected PDF components: (p_ph_sig)
[#1] INFO:Plotting -- RooAbsPdf::plotOn(model1) indirectly selected PDF components: (sp_ph)
[#1] INFO:Plotting -- RooAbsPdf::plotOn(model1) directly selected PDF components: (p_ph_bkg)
[#1] INFO:Plotting -- RooAbsPdf::plotOn(model1) indirectly selected PDF components: (sp_ph)
[#1] INFO:Plotting -- RooPlot::updateFitRangeNorm: New event count of 50 will supercede previous event count of 1050 for normalization of PDF projections
[#1] INFO:Plotting -- RooAbsPdf::plotOn(model2) directly selected PDF components: (p_ph_sig2)
[#1] INFO:Plotting -- RooAbsPdf::plotOn(model2) indirectly selected PDF components: (sp_ph)
[#1] INFO:Plotting -- RooAbsPdf::plotOn(model2) directly selected PDF components: (p_ph_bkg2)
[#1] INFO:Plotting -- RooAbsPdf::plotOn(model2) indirectly selected PDF components: (sp_ph)
[#1] INFO:Plotting -- RooPlot::updateFitRangeNorm: New event count of 50 will supercede previous event count of 1050 for normalization of PDF projections
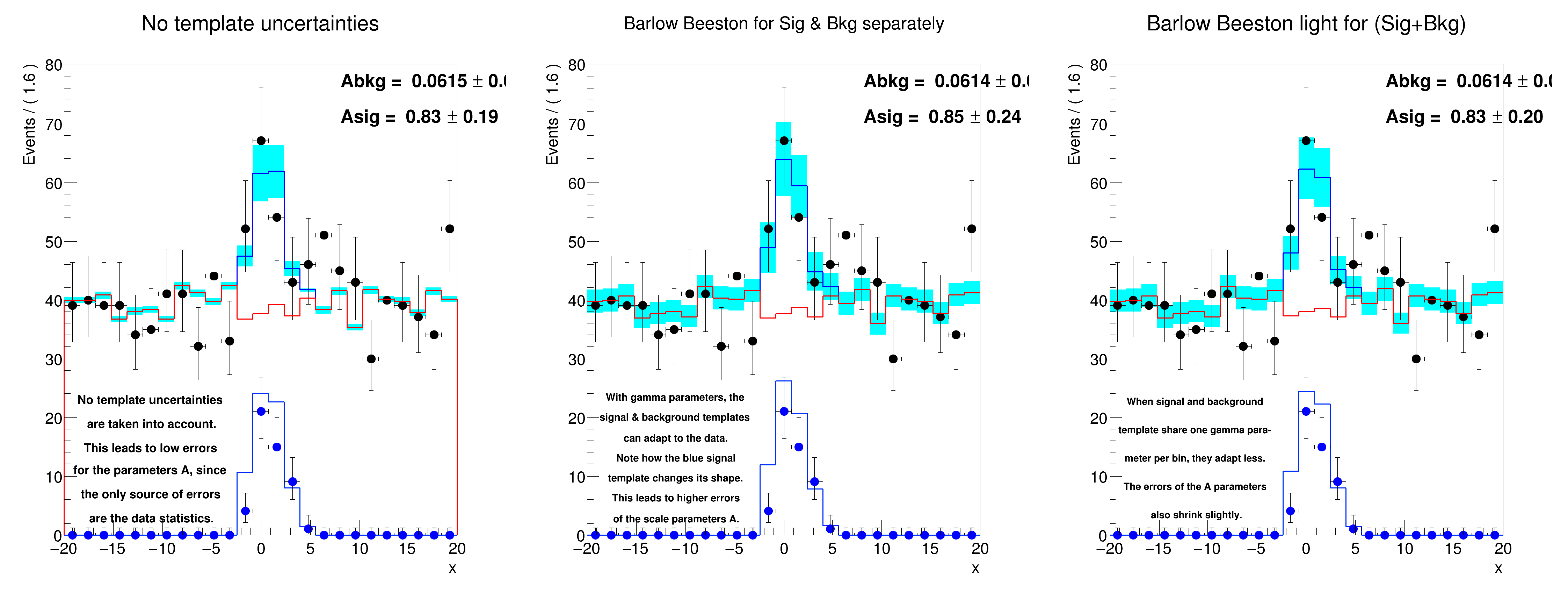
Asig [normal ] = 0.8337777298752981 +/- 0.18981418231893016
Asig [BB ] = 0.8505069710009863 +/- 0.2357777227529776
Asig [BBlight] = 0.8344080864402419 +/- 0.2029724823213735
import ROOT
x = ROOT.RooRealVar("x", "x", -20, 20)
x.setBins(25)
meanG = ROOT.RooRealVar("meanG", "meanG", 1, -10, 10)
sigG = ROOT.RooRealVar("sigG", "sigG", 1.5, -10, 10)
g = ROOT.RooGaussian("g", "Gauss", x, meanG, sigG)
u = ROOT.RooUniform("u", "Uniform", x)
sigData = g.generate(x, 50)
bkgData = u.generate(x, 1000)
sumData = ROOT.RooDataSet("sumData", "Gauss + Uniform", x)
sumData.append(sigData)
sumData.append(bkgData)
dh_sig = g.generateBinned(x, 50)
dh_bkg = u.generateBinned(x, 10000)
p_h_sig = ROOT.RooHistFunc("p_h_sig", "p_h_sig", x, dh_sig)
p_h_bkg = ROOT.RooHistFunc("p_h_bkg", "p_h_bkg", x, dh_bkg)
Asig0 = ROOT.RooRealVar("Asig", "Asig", 1, 0.01, 5000)
Abkg0 = ROOT.RooRealVar("Abkg", "Abkg", 1, 0.01, 5000)
model0 = ROOT.RooRealSumPdf("model0", "model0", [p_h_sig, p_h_bkg], [Asig0, Abkg0], True)
p_ph_sig1 = ROOT.RooParamHistFunc("p_ph_sig", "p_ph_sig", dh_sig)
p_ph_bkg1 = ROOT.RooParamHistFunc("p_ph_bkg", "p_ph_bkg", dh_bkg)
Asig1 = ROOT.RooRealVar("Asig", "Asig", 1, 0.01, 5000)
Abkg1 = ROOT.RooRealVar("Abkg", "Abkg", 1, 0.01, 5000)
model_tmp = ROOT.RooRealSumPdf("sp_ph", "sp_ph", [p_ph_sig1, p_ph_bkg1], [Asig1, Abkg1], True)
hc_sig = ROOT.RooHistConstraint("hc_sig", "hc_sig", p_ph_sig1)
hc_bkg = ROOT.RooHistConstraint("hc_bkg", "hc_bkg", p_ph_bkg1)
model1 = ROOT.RooProdPdf("model1", "model1", {hc_sig, hc_bkg}, Conditional=(model_tmp, x))
p_ph_sig2 = ROOT.RooParamHistFunc("p_ph_sig2", "p_ph_sig2", dh_sig)
p_ph_bkg2 = ROOT.RooParamHistFunc("p_ph_bkg2", "p_ph_bkg2", dh_bkg, p_ph_sig2, True)
Asig2 = ROOT.RooRealVar("Asig", "Asig", 1, 0.01, 5000)
Abkg2 = ROOT.RooRealVar("Abkg", "Abkg", 1, 0.01, 5000)
model2_tmp = ROOT.RooRealSumPdf("sp_ph", "sp_ph", [p_ph_sig2, p_ph_bkg2], [Asig2, Abkg2], True)
hc_sigbkg = ROOT.RooHistConstraint("hc_sigbkg", "hc_sigbkg", {p_ph_sig2, p_ph_bkg2})
model2 = ROOT.RooProdPdf("model2", "model2", hc_sigbkg, Conditional=(model2_tmp, x))
result0 = model0.fitTo(sumData, PrintLevel=0, Save=True)
result1 = model1.fitTo(sumData, PrintLevel=0, Save=True)
result2 = model2.fitTo(sumData, PrintLevel=0, Save=True)
can = ROOT.TCanvas("can", "", 1500, 600)
can.Divide(3, 1)
pt = ROOT.TPaveText(-19.5, 1, -2, 25)
pt.SetFillStyle(0)
pt.SetBorderSize(0)
can.cd(1)
frame = x.frame(Title="No template uncertainties")
sumData.plotOn(frame)
model0.plotOn(frame, LineColor="b", VisualizeError=result0)
sumData.plotOn(frame)
model0.plotOn(frame, LineColor="b")
p_ph_sig_set = {p_h_sig}
p_ph_bkg_set = {p_h_bkg}
model0.plotOn(frame, Components=p_ph_sig_set, LineColor="kAzure")
model0.plotOn(frame, Components=p_ph_bkg_set, LineColor="r")
model0.paramOn(frame)
sigData.plotOn(frame, MarkerColor="b")
frame.Draw()
pt_text1 = [
"No template uncertainties",
"are taken into account.",
"This leads to low errors",
"for the parameters A, since",
"the only source of errors",
"are the data statistics.",
]
for text in pt_text1:
pt.AddText(text)
pt.DrawClone()
can.cd(2)
frame = x.frame(Title="Barlow Beeston for Sig & Bkg separately")
sumData.plotOn(frame)
model1.plotOn(frame, LineColor="b", VisualizeError=result1)
sumData.plotOn(frame)
model1.plotOn(frame, LineColor="b")
p_ph_sig1_set = {p_ph_sig1}
p_ph_bkg1_set = {p_ph_bkg1}
model1.plotOn(frame, Components=p_ph_sig1_set, LineColor="kAzure")
model1.plotOn(frame, Components=p_ph_bkg1_set, LineColor="r")
model1.paramOn(frame, Parameters={Asig1, Abkg1})
sigData.plotOn(frame, MarkerColor="b")
frame.Draw()
pt.Clear()
pt_text2 = [
"With gamma parameters, the",
"signal & background templates",
"can adapt to the data.",
"Note how the blue signal",
"template changes its shape.",
"This leads to higher errors",
"of the scale parameters A.",
]
for text in pt_text2:
pt.AddText(text)
pt.DrawClone()
can.cd(3)
frame = x.frame(Title="Barlow Beeston light for (Sig+Bkg)")
sumData.plotOn(frame)
model2.plotOn(frame, LineColor="b", VisualizeError=result2)
sumData.plotOn(frame)
model2.plotOn(frame, LineColor="b")
p_ph_sig2_set = {p_ph_sig2}
p_ph_bkg2_set = {p_ph_bkg2}
model2.plotOn(frame, Components=p_ph_sig2_set, LineColor="kAzure")
model2.plotOn(frame, Components=p_ph_bkg2_set, LineColor="r")
model2.paramOn(frame, Parameters={Asig2, Abkg2})
sigData.plotOn(frame, MarkerColor="b")
frame.Draw()
pt.Clear()
pt_text3 = [
"When signal and background",
"template share one gamma para-",
"meter per bin, they adapt less.",
"The errors of the A parameters",
"also shrink slightly.",
]
for text in pt_text3:
pt.AddText(text)
pt.DrawClone()
print("Asig [normal ] = {} +/- {}".format(Asig0.getVal(), Asig0.getError()))
print("Asig [BB ] = {} +/- {}".format(Asig1.getVal(), Asig1.getError()))
print("Asig [BBlight] = {} +/- {}".format(Asig2.getVal(), Asig2.getError()))
can.SaveAs("rf709_BarlowBeeston.png")

 Implementing the Barlow-Beeston method for taking into account the statistical uncertainty of a Monte-Carlo fit template.
Implementing the Barlow-Beeston method for taking into account the statistical uncertainty of a Monte-Carlo fit template.