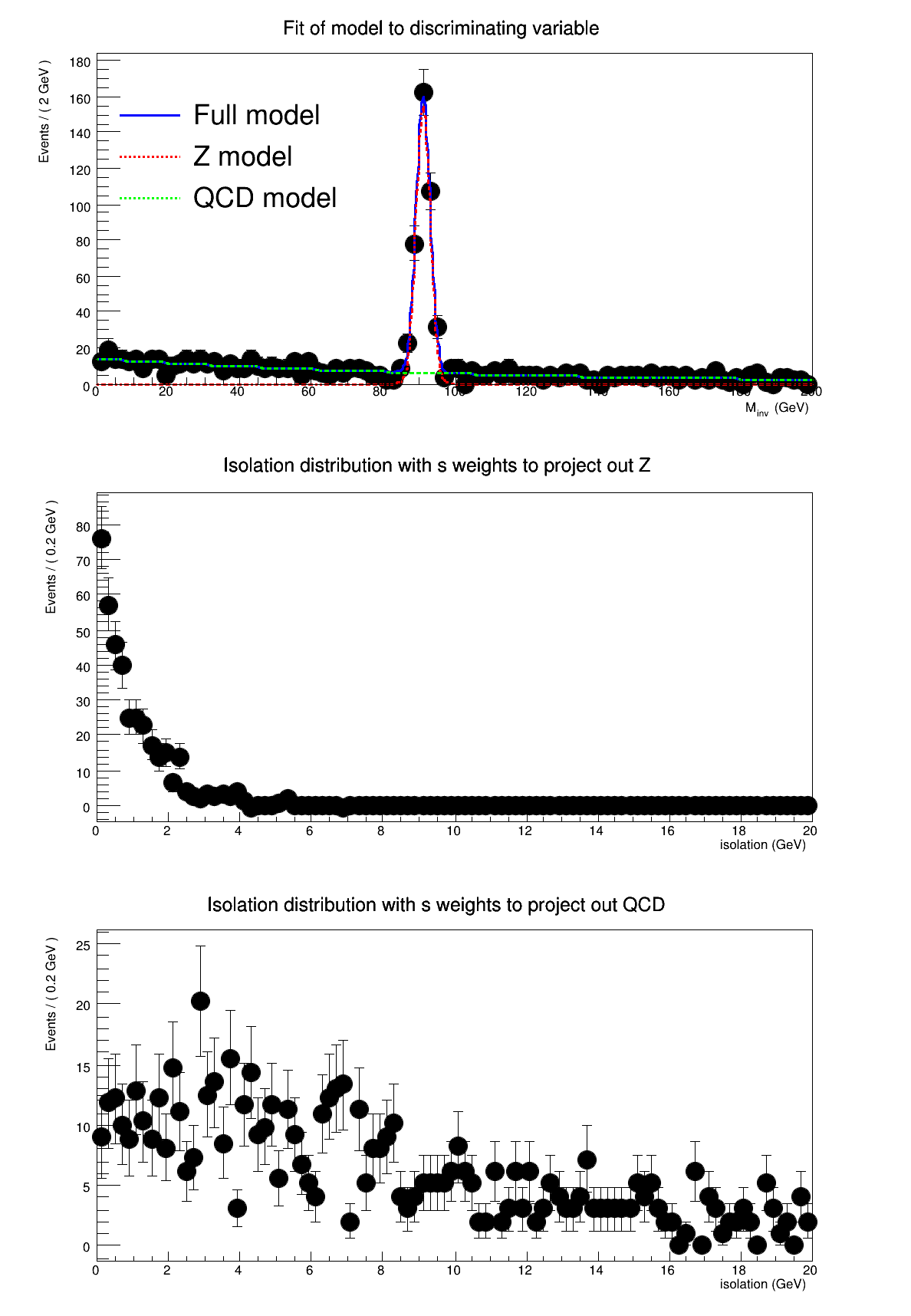
This tutorial shows an example of using SPlot to unfold two distributions. The physics context for the example is that we want to know the isolation distribution for real electrons from Z events and fake electrons from QCD. Isolation is our 'control' variable. To unfold them, we need a model for an uncorrelated variable that discriminates between Z and QCD. To do this, we use the invariant mass of two electrons. We model the Z with a Gaussian and the QCD with a falling exponential.
Note, since we don't have real data in this tutorial, we need to generate toy data. To do that we need a model for the isolation variable for both Z and QCD. This is only used to generate the toy data, and would not be needed if we had real data.
#include <iomanip>
void rs301_splot()
{
AddModel(wspace);
AddData(wspace);
DoSPlot(wspace);
MakePlots(wspace);
delete wspace;
}
{
Double_t lowRange = 0., highRange = 200.;
RooRealVar invMass(
"invMass",
"M_{inv}", lowRange, highRange,
"GeV");
RooRealVar isolation(
"isolation",
"isolation", 0., 20.,
"GeV");
std::cout << "make z model" << std::endl;
RooRealVar mZ(
"mZ",
"Z Mass", 91.2, lowRange, highRange);
RooRealVar sigmaZ(
"sigmaZ",
"Width of Gaussian", 2, 0, 10,
"GeV");
RooGaussian mZModel(
"mZModel",
"Z+jets Model", invMass, mZ, sigmaZ);
mZ.setConstant();
RooConstVar zIsolDecayConst(
"zIsolDecayConst",
"z isolation decay constant", -1);
RooExponential zIsolationModel(
"zIsolationModel",
"z isolation model", isolation, zIsolDecayConst);
std::cout << "make qcd model" << std::endl;
RooRealVar qcdMassDecayConst(
"qcdMassDecayConst",
"Decay const for QCD mass spectrum", -0.01, -100, 100,
"1/GeV");
RooExponential qcdMassModel(
"qcdMassModel",
"qcd Mass Model", invMass, qcdMassDecayConst);
RooConstVar qcdIsolDecayConst(
"qcdIsolDecayConst",
"Et resolution constant", -.1);
RooExponential qcdIsolationModel(
"qcdIsolationModel",
"QCD isolation model", isolation, qcdIsolDecayConst);
RooProdPdf qcdModel(
"qcdModel",
"2-d model for QCD",
RooArgSet(qcdMassModel, qcdIsolationModel));
RooRealVar zYield(
"zYield",
"fitted yield for Z", 50, 0., 1000);
RooRealVar qcdYield(
"qcdYield",
"fitted yield for QCD", 100, 0., 1000);
std::cout << "make full model" << std::endl;
model.graphVizTree("fullModel.dot");
std::cout << "import model" << std::endl;
}
{
std::cout << "make data set and import to workspace" << std::endl;
}
{
std::cout << "Calculate sWeights" << std::endl;
std::cout << "\n\n------------------------------------------\nThe dataset before creating sWeights:\n";
std::cout << "\n\nThe dataset after creating sWeights:\n";
std::cout << "\n\n------------------------------------------\n\nCheck SWeights:" << std::endl;
std::cout << std::endl
<<
"Yield of Z is\t" << zYield->
getVal() <<
". From sWeights it is "
std::cout <<
"Yield of QCD is\t" << qcdYield->
getVal() <<
". From sWeights it is " << std::endl;
for (
Int_t i = 0; i < 10; i++) {
std::cout <<
"z Weight for event " << i << std::right << std::setw(12) << sData->
GetSWeight(i,
"zYield") <<
" qcd Weight" << std::endl;
}
std::cout << std::endl;
std::cout << "import new dataset with sWeights" << std::endl;
ws->import(*data,
Rename(
"dataWithSWeights"));
}
{
std::cout << "make plots" << std::endl;
frame->
SetTitle(
"Fit of model to discriminating variable");
frame2->
SetTitle(
"Isolation distribution with s weights to project out Z");
frame3->
SetTitle(
"Isolation distribution with s weights to project out QCD");
}
virtual void Print(Option_t *options=0) const
Print TNamed name and title.
virtual RooPlot * plotOn(RooPlot *frame, const RooCmdArg &arg1=RooCmdArg::none(), const RooCmdArg &arg2=RooCmdArg::none(), const RooCmdArg &arg3=RooCmdArg::none(), const RooCmdArg &arg4=RooCmdArg::none(), const RooCmdArg &arg5=RooCmdArg::none(), const RooCmdArg &arg6=RooCmdArg::none(), const RooCmdArg &arg7=RooCmdArg::none(), const RooCmdArg &arg8=RooCmdArg::none()) const
RooDataSet * generate(const RooArgSet &whatVars, Int_t nEvents, const RooCmdArg &arg1, const RooCmdArg &arg2=RooCmdArg::none(), const RooCmdArg &arg3=RooCmdArg::none(), const RooCmdArg &arg4=RooCmdArg::none(), const RooCmdArg &arg5=RooCmdArg::none())
See RooAbsPdf::generate(const RooArgSet&,const RooCmdArg&,const RooCmdArg&,const RooCmdArg&,...
virtual RooFitResult * fitTo(RooAbsData &data, const RooCmdArg &arg1=RooCmdArg::none(), const RooCmdArg &arg2=RooCmdArg::none(), const RooCmdArg &arg3=RooCmdArg::none(), const RooCmdArg &arg4=RooCmdArg::none(), const RooCmdArg &arg5=RooCmdArg::none(), const RooCmdArg &arg6=RooCmdArg::none(), const RooCmdArg &arg7=RooCmdArg::none(), const RooCmdArg &arg8=RooCmdArg::none())
Fit PDF to given dataset.
virtual RooPlot * plotOn(RooPlot *frame, const RooCmdArg &arg1=RooCmdArg::none(), const RooCmdArg &arg2=RooCmdArg::none(), const RooCmdArg &arg3=RooCmdArg::none(), const RooCmdArg &arg4=RooCmdArg::none(), const RooCmdArg &arg5=RooCmdArg::none(), const RooCmdArg &arg6=RooCmdArg::none(), const RooCmdArg &arg7=RooCmdArg::none(), const RooCmdArg &arg8=RooCmdArg::none(), const RooCmdArg &arg9=RooCmdArg::none(), const RooCmdArg &arg10=RooCmdArg::none()) const
Helper calling plotOn(RooPlot*, RooLinkedList&) const.
RooPlot * frame(const RooCmdArg &arg1, const RooCmdArg &arg2=RooCmdArg::none(), const RooCmdArg &arg3=RooCmdArg::none(), const RooCmdArg &arg4=RooCmdArg::none(), const RooCmdArg &arg5=RooCmdArg::none(), const RooCmdArg &arg6=RooCmdArg::none(), const RooCmdArg &arg7=RooCmdArg::none(), const RooCmdArg &arg8=RooCmdArg::none()) const
Create a new RooPlot on the heap with a drawing frame initialized for this object,...
Double_t getVal(const RooArgSet *normalisationSet=nullptr) const
Evaluate object.
RooAddPdf is an efficient implementation of a sum of PDFs of the form.
RooArgList is a container object that can hold multiple RooAbsArg objects.
RooArgSet is a container object that can hold multiple RooAbsArg objects.
RooConstVar represent a constant real-valued object.
RooDataSet is a container class to hold unbinned data.
virtual const RooArgSet * get(Int_t index) const override
Return RooArgSet with coordinates of event 'index'.
static RooMsgService & instance()
Return reference to singleton instance.
void setGlobalKillBelow(RooFit::MsgLevel level)
void setSilentMode(Bool_t flag)
A RooPlot is a plot frame and a container for graphics objects within that frame.
void SetTitle(const char *name)
Set the title of the RooPlot to 'title'.
TObject * findObject(const char *name, const TClass *clas=0) const
Find the named object in our list of items and return a pointer to it.
virtual void Draw(Option_t *options=0)
Draw this plot and all of the elements it contains.
RooProdPdf is an efficient implementation of a product of PDFs of the form.
RooRealVar represents a variable that can be changed from the outside.
A class to calculate "sWeights" used to create an "sPlot".
Double_t GetYieldFromSWeight(const char *sVariable) const
Sum the SWeights for a particular species over all events.
Double_t GetSWeight(Int_t numEvent, const char *sVariable) const
Retrieve an s weight.
Double_t GetSumOfEventSWeight(Int_t numEvent) const
Sum the SWeights for a particular event.
The RooWorkspace is a persistable container for RooFit projects.
TVirtualPad * cd(Int_t subpadnumber=0) override
Set current canvas & pad.
This class displays a legend box (TPaveText) containing several legend entries.
virtual const char * GetTitle() const
Returns title of object.
virtual const char * GetName() const
Returns name of object.
void Divide(Int_t nx=1, Int_t ny=1, Float_t xmargin=0.01, Float_t ymargin=0.01, Int_t color=0) override
Automatic pad generation by division.
RooCmdArg Rename(const char *suffix)
RooCmdArg Extended(Bool_t flag=kTRUE)
RooCmdArg DataError(Int_t)
RooCmdArg Components(const RooArgSet &compSet)
RooCmdArg LineColor(Color_t color)
RooCmdArg LineStyle(Style_t style)
The namespace RooFit contains mostly switches that change the behaviour of functions of PDFs (or othe...
Namespace for the RooStats classes.
␛[1mRooFit v3.60 -- Developed by Wouter Verkerke and David Kirkby␛[0m
Copyright (C) 2000-2013 NIKHEF, University of California & Stanford University
All rights reserved, please read http://roofit.sourceforge.net/license.txt
make z model
[#0] WARNING:InputArguments -- The parameter 'sigmaZ' with range [0, 10] of the RooGaussian 'mZModel' exceeds the safe range of (0, inf). Advise to limit its range.
make qcd model
make full model
import model
[#1] INFO:ObjectHandling -- RooWorkspace::import(myWS) importing RooAddPdf::model
[#1] INFO:ObjectHandling -- RooWorkspace::import(myWS) importing RooProdPdf::zModel
[#1] INFO:ObjectHandling -- RooWorkspace::import(myWS) importing RooGaussian::mZModel
[#1] INFO:ObjectHandling -- RooWorkspace::import(myWS) importing RooRealVar::invMass
[#1] INFO:ObjectHandling -- RooWorkspace::import(myWS) importing RooRealVar::mZ
[#1] INFO:ObjectHandling -- RooWorkspace::import(myWS) importing RooRealVar::sigmaZ
[#1] INFO:ObjectHandling -- RooWorkspace::import(myWS) importing RooExponential::zIsolationModel
[#1] INFO:ObjectHandling -- RooWorkspace::import(myWS) importing RooRealVar::isolation
[#1] INFO:ObjectHandling -- RooWorkspace::import(myWS) importing RooConstVar::zIsolDecayConst
[#1] INFO:ObjectHandling -- RooWorkspace::import(myWS) importing RooRealVar::zYield
[#1] INFO:ObjectHandling -- RooWorkspace::import(myWS) importing RooProdPdf::qcdModel
[#1] INFO:ObjectHandling -- RooWorkspace::import(myWS) importing RooExponential::qcdMassModel
[#1] INFO:ObjectHandling -- RooWorkspace::import(myWS) importing RooRealVar::qcdMassDecayConst
[#1] INFO:ObjectHandling -- RooWorkspace::import(myWS) importing RooExponential::qcdIsolationModel
[#1] INFO:ObjectHandling -- RooWorkspace::import(myWS) importing RooConstVar::qcdIsolDecayConst
[#1] INFO:ObjectHandling -- RooWorkspace::import(myWS) importing RooRealVar::qcdYield
make data set and import to workspace
[#1] INFO:ObjectHandling -- RooWorkspace::import(myWS) importing dataset modelData
[#1] INFO:ObjectHandling -- RooWorkSpace::import(myWS) changing name of dataset from modelData to data
Calculate sWeights
[#1] INFO:Minization -- createNLL: caching constraint set under name CONSTR_OF_PDF_model_FOR_OBS_invMass:isolation with 0 entries
[#1] INFO:Minization -- RooMinimizer::optimizeConst: activating const optimization
[#1] INFO:Minization -- The following expressions have been identified as constant and will be precalculated and cached: (zIsolationModel,qcdIsolationModel)
[#1] INFO:Minization -- The following expressions will be evaluated in cache-and-track mode: (mZModel,qcdMassModel)
**********
** 1 **SET PRINT 1
**********
**********
** 2 **SET NOGRAD
**********
PARAMETER DEFINITIONS:
NO. NAME VALUE STEP SIZE LIMITS
1 qcdMassDecayConst -1.00000e-02 2.00000e+01 -1.00000e+02 1.00000e+02
2 qcdYield 1.00000e+02 5.00000e+01 0.00000e+00 1.00000e+03
3 sigmaZ 2.00000e+00 1.00000e+00 0.00000e+00 1.00000e+01
4 zYield 5.00000e+01 2.50000e+01 0.00000e+00 1.00000e+03
**********
** 3 **SET ERR 0.5
**********
**********
** 4 **SET PRINT 1
**********
**********
** 5 **SET STR 1
**********
NOW USING STRATEGY 1: TRY TO BALANCE SPEED AGAINST RELIABILITY
**********
** 6 **MIGRAD 2000 1
**********
FIRST CALL TO USER FUNCTION AT NEW START POINT, WITH IFLAG=4.
START MIGRAD MINIMIZATION. STRATEGY 1. CONVERGENCE WHEN EDM .LT. 1.00e-03
FCN=1901.86 FROM MIGRAD STATUS=INITIATE 18 CALLS 19 TOTAL
EDM= unknown STRATEGY= 1 NO ERROR MATRIX
EXT PARAMETER CURRENT GUESS STEP FIRST
NO. NAME VALUE ERROR SIZE DERIVATIVE
1 qcdMassDecayConst -1.00000e-02 2.00000e+01 2.01358e-01 -1.06814e+05
2 qcdYield 1.00000e+02 5.00000e+01 1.72186e-01 -1.57317e+03
3 sigmaZ 2.00000e+00 1.00000e+00 2.57889e-01 3.62916e+01
4 zYield 5.00000e+01 2.50000e+01 1.18625e-01 -1.41930e+03
ERR DEF= 0.5
MIGRAD MINIMIZATION HAS CONVERGED.
MIGRAD WILL VERIFY CONVERGENCE AND ERROR MATRIX.
COVARIANCE MATRIX CALCULATED SUCCESSFULLY
FCN=849.923 FROM MIGRAD STATUS=CONVERGED 110 CALLS 111 TOTAL
EDM=3.1409e-06 STRATEGY= 1 ERROR MATRIX ACCURATE
EXT PARAMETER STEP FIRST
NO. NAME VALUE ERROR SIZE DERIVATIVE
1 qcdMassDecayConst -9.30176e-03 7.55562e-04 1.52115e-07 2.51004e+01
2 qcdYield 6.22140e+02 2.52789e+01 1.04861e-03 -1.45735e-02
3 sigmaZ 1.95257e+00 8.09927e-02 4.09416e-04 -7.77193e-02
4 zYield 3.77836e+02 1.98770e+01 8.24140e-04 -6.51095e-03
ERR DEF= 0.5
EXTERNAL ERROR MATRIX. NDIM= 25 NPAR= 4 ERR DEF=0.5
5.709e-07 1.999e-04 -1.007e-06 -1.999e-04
1.999e-04 6.396e+02 -9.256e-02 -1.747e+01
-1.007e-06 -9.256e-02 6.561e-03 9.257e-02
-1.999e-04 -1.747e+01 9.257e-02 3.953e+02
PARAMETER CORRELATION COEFFICIENTS
NO. GLOBAL 1 2 3 4
1 0.02260 1.000 0.010 -0.016 -0.013
2 0.05626 0.010 1.000 -0.045 -0.035
3 0.07351 -0.016 -0.045 1.000 0.057
4 0.06697 -0.013 -0.035 0.057 1.000
**********
** 7 **SET ERR 0.5
**********
**********
** 8 **SET PRINT 1
**********
**********
** 9 **HESSE 2000
**********
COVARIANCE MATRIX CALCULATED SUCCESSFULLY
FCN=849.923 FROM HESSE STATUS=OK 23 CALLS 134 TOTAL
EDM=3.13931e-06 STRATEGY= 1 ERROR MATRIX ACCURATE
EXT PARAMETER INTERNAL INTERNAL
NO. NAME VALUE ERROR STEP SIZE VALUE
1 qcdMassDecayConst -9.30176e-03 7.55563e-04 3.04229e-08 -9.30176e-05
2 qcdYield 6.22140e+02 2.52790e+01 2.09721e-04 2.46778e-01
3 sigmaZ 1.95257e+00 8.09935e-02 8.18832e-05 -6.55413e-01
4 zYield 3.77836e+02 1.98772e+01 1.64828e-04 -2.46827e-01
ERR DEF= 0.5
EXTERNAL ERROR MATRIX. NDIM= 25 NPAR= 4 ERR DEF=0.5
5.709e-07 2.002e-04 -1.009e-06 -2.002e-04
2.002e-04 6.396e+02 -9.273e-02 -1.749e+01
-1.009e-06 -9.273e-02 6.561e-03 9.273e-02
-2.002e-04 -1.749e+01 9.273e-02 3.953e+02
PARAMETER CORRELATION COEFFICIENTS
NO. GLOBAL 1 2 3 4
1 0.02264 1.000 0.010 -0.016 -0.013
2 0.05634 0.010 1.000 -0.045 -0.035
3 0.07364 -0.016 -0.045 1.000 0.058
4 0.06707 -0.013 -0.035 0.058 1.000
[#1] INFO:Minization -- RooMinimizer::optimizeConst: deactivating const optimization
------------------------------------------
The dataset before creating sWeights:
RooDataSet::data[invMass,isolation] = 1000 entries
The dataset after creating sWeights:
RooDataSet::data[invMass,isolation,zYield_sw,L_zYield,qcdYield_sw,L_qcdYield] = 1000 entries
------------------------------------------
Check SWeights:
Yield of Z is 377.841. From sWeights it is 377.841
Yield of QCD is 622.16. From sWeights it is 622.16
z Weight for event 0 -0.0259121 qcd Weight 1.02591 Total Weight 1
z Weight for event 1 -0.0259121 qcd Weight 1.02591 Total Weight 1
z Weight for event 2 1.02602 qcd Weight -0.0260232 Total Weight 1
z Weight for event 3 1.01358 qcd Weight -0.0135828 Total Weight 1
z Weight for event 4 -0.0259121 qcd Weight 1.02591 Total Weight 1
z Weight for event 5 1.02684 qcd Weight -0.0268354 Total Weight 1
z Weight for event 6 -0.0259121 qcd Weight 1.02591 Total Weight 1
z Weight for event 7 -0.0259121 qcd Weight 1.02591 Total Weight 1
z Weight for event 8 1.03438 qcd Weight -0.0343802 Total Weight 1
z Weight for event 9 0.910182 qcd Weight 0.0898196 Total Weight 1
import new dataset with sWeights
make plots
[#1] INFO:Plotting -- RooAbsReal::plotOn(model) plot on invMass integrates over variables (isolation)
[#1] INFO:Plotting -- RooAbsPdf::plotOn(model) directly selected PDF components: (zModel)
[#1] INFO:Plotting -- RooAbsPdf::plotOn(model) indirectly selected PDF components: (mZModel,zIsolationModel)
[#1] INFO:Plotting -- RooAbsReal::plotOn(model) plot on invMass integrates over variables (isolation)
[#1] INFO:Plotting -- RooAbsPdf::plotOn(model) directly selected PDF components: (qcdModel)
[#1] INFO:Plotting -- RooAbsPdf::plotOn(model) indirectly selected PDF components: (qcdMassModel,qcdIsolationModel)
[#1] INFO:Plotting -- RooAbsReal::plotOn(model) plot on invMass integrates over variables (isolation)