␛[1mRooFit v3.60 -- Developed by Wouter Verkerke and David Kirkby␛[0m
Copyright (C) 2000-2013 NIKHEF, University of California & Stanford University
All rights reserved, please read http://roofit.sourceforge.net/license.txt
DataStore d (d)
Contains 1000 entries
Observables:
1) x = 9 L(-10 - 10) "x"
2) y = 31.607 L(0 - 40) "y"
3) c = Plus(idx = 1)
"c"
1) 0x55e550426020 RooRealVar:: x = 9 L(-10 - 10) "x"
2) 0x55e550796a50 RooRealVar:: y = 31.607 L(0 - 40) "y"
3) 0x55e550772440 RooCategory:: c = Plus(idx = 1)
"c"
1) 0x55e550426020 RooRealVar:: x = 8 L(-10 - 10) "x"
2) 0x55e550796a50 RooRealVar:: y = 30 L(0 - 40) "y"
3) 0x55e550772440 RooCategory:: c = Minus(idx = -1)
"c"
>> d1 has only columns x,c
DataStore d (d)
Contains 1000 entries
Observables:
1) x = 9 L(-10 - 10) "x"
2) c = Plus(idx = 1)
"c"
>> d2 has only column y
DataStore d (d)
Contains 1000 entries
Observables:
1) y = 31.607 L(0 - 40) "y"
>> d3 has only the points with y>5.17
DataStore d (d)
Contains 973 entries
Observables:
1) x = 9 L(-10 - 10) "x"
2) y = 31.607 L(0 - 40) "y"
3) c = Plus(idx = 1)
"c"
>> d4 has only columns x,c for data points with y>5.17
DataStore d (d)
Contains 973 entries
Observables:
1) x = 9 L(-10 - 10) "x"
2) c = Plus(idx = 1)
"c"
>> merge d2(y) with d1(x,c) to form d1(x,c,y)
DataStore d (d)
Contains 1000 entries
Observables:
1) x = 9 L(-10 - 10) "x"
2) c = Plus(idx = 1)
"c"
3) y = 31.607 L(0 - 40) "y"
>> append data points of d3 to d1
DataStore d (d)
Contains 1973 entries
Observables:
1) x = 9 L(-10 - 10) "x"
2) c = Plus(idx = 1)
"c"
3) y = 31.607 L(0 - 40) "y"
>> construct dh (binned) from d(unbinned) but only take the x and y dimensions,
>> the category 'c' will be projected in the filling process
DataStore dh (binned version of d)
Contains 100 entries
Observables:
1) x = 9 L(-10 - 10) B(10) "x"
2) y = 31.607 L(0 - 40) B(10) "y"
Binned Dataset dh (binned version of d)
Contains 100 bins with a total weight of 1000
Observables: 1) x = 9 L(-10 - 10) B(10) "x"
2) y = 31.607 L(0 - 40) B(10) "y"
>> number of bins in dh : 100
>> sum of weights in dh : 1000
>> integral over histogram: 8000
>> retrieving the properties of the bin enclosing coordinate (x,y) = (0.3,20.5)
bin center:
1) 0x55e55059ef60 RooRealVar:: x = 1 L(-10 - 10) B(10) "x"
2) 0x55e55087c170 RooRealVar:: y = 22 L(0 - 40) B(10) "y"
weight = 76
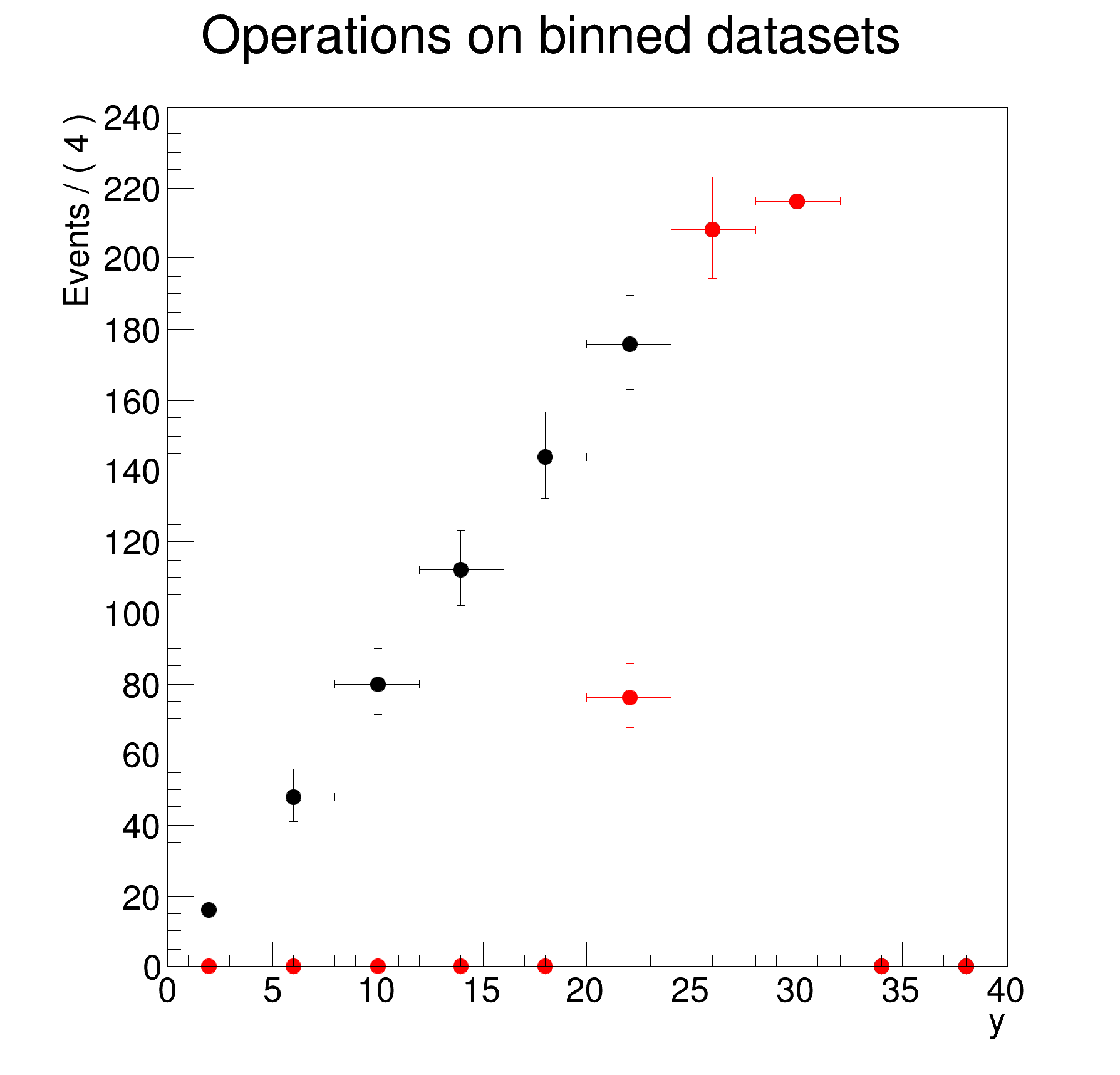
>> Creating 1-dimensional projection on y of dh for bins with x>0
DataStore dh (binned version of d)
Contains 10 entries
Observables:
1) y = 38 L(0 - 40) B(10) "y"
Binned Dataset dh (binned version of d)
Contains 10 bins with a total weight of 500
Observables: 1) y = 38 L(0 - 40) B(10) "y"
[#1] INFO:Plotting -- RooPlot::updateFitRangeNorm: New event count of 500 will supercede previous event count of 1000 for normalization of PDF projections
>> Persisting d via ROOT I/O
TFile** rf402_datahandling.root
TFile* rf402_datahandling.root
KEY: RooDataSet d;1 d
KEY: TProcessID ProcessID0;1 c9599a9c-bac5-11ec-b0d2-942c8a89beef
{
c.defineType(
"Plus", +1);
c.defineType(
"Minus", -1);
for (i = 0; i < 1000; i++) {
c.setLabel((i % 2) ?
"Plus" :
"Minus");
}
cout << endl;
cout << endl;
cout << endl;
cout << endl << ">> d1 has only columns x,c" << endl;
cout << endl << ">> d2 has only column y" << endl;
cout << endl << ">> d3 has only the points with y>5.17" << endl;
cout << endl << ">> d4 has only columns x,c for data points with y>5.17" << endl;
cout << endl << ">> merge d2(y) with d1(x,c) to form d1(x,c,y)" << endl;
cout << endl << ">> append data points of d3 to d1" << endl;
cout << ">> construct dh (binned) from d(unbinned) but only take the x and y dimensions," << endl
<< ">> the category 'c' will be projected in the filling process" << endl;
dh.Print("v");
dh.plotOn(yframe);
cout << ">> number of bins in dh : " << dh.numEntries() << endl;
cout <<
">> sum of weights in dh : " << dh.sum(
kFALSE) << endl;
cout <<
">> integral over histogram: " << dh.sum(
kTRUE) << endl;
cout << ">> retrieving the properties of the bin enclosing coordinate (x,y) = (0.3,20.5) " << endl;
cout << " bin center:" << endl;
cout << " weight = " << dh.weight() << endl;
cout << ">> Creating 1-dimensional projection on y of dh for bins with x>0" << endl;
cout << endl << ">> Persisting d via ROOT I/O" << endl;
TFile f(
"rf402_datahandling.root",
"RECREATE");
new TCanvas(
"rf402_datahandling",
"rf402_datahandling", 600, 600);
gPad->SetLeftMargin(0.15);
}
virtual void Print(Option_t *options=0) const
This method must be overridden when a class wants to print itself.
RooAbsData * reduce(const RooCmdArg &arg1, const RooCmdArg &arg2=RooCmdArg(), const RooCmdArg &arg3=RooCmdArg(), const RooCmdArg &arg4=RooCmdArg(), const RooCmdArg &arg5=RooCmdArg(), const RooCmdArg &arg6=RooCmdArg(), const RooCmdArg &arg7=RooCmdArg(), const RooCmdArg &arg8=RooCmdArg())
Create a reduced copy of this dataset.
virtual void Print(Option_t *options=0) const
Print TNamed name and title.
RooArgSet is a container object that can hold multiple RooAbsArg objects.
RooCategory represents a fundamental (non-derived) discrete value object.
The RooDataHist is a container class to hold N-dimensional binned data.
virtual RooPlot * plotOn(RooPlot *frame, PlotOpt o) const
Back end function to plotting functionality.
RooDataSet is a container class to hold unbinned data.
void append(RooDataSet &data)
Add all data points of given data set to this data set.
Bool_t merge(RooDataSet *data1, RooDataSet *data2=0, RooDataSet *data3=0, RooDataSet *data4=0, RooDataSet *data5=0, RooDataSet *data6=0)
A RooPlot is a plot frame and a container for graphics objects within that frame.
virtual void Draw(Option_t *options=0)
Draw this plot and all of the elements it contains.
RooRealVar represents a fundamental (non-derived) real valued object.
virtual void SetTitleOffset(Float_t offset=1)
Set distance between the axis and the axis title Offset is a correction factor with respect to the "s...
A ROOT file is a suite of consecutive data records (TKey instances) with a well defined format.
Template specialisation used in RooAbsArg:
RooCmdArg MarkerColor(Color_t color)
RooCmdArg LineColor(Color_t color)
RooCmdArg Bins(Int_t nbin)



 Data and categories: tools for manipulation of (un)binned datasets
Data and categories: tools for manipulation of (un)binned datasets