␛[1mRooFit v3.60 -- Developed by Wouter Verkerke and David Kirkby␛[0m
Copyright (C) 2000-2013 NIKHEF, University of California & Stanford University
All rights reserved, please read http://roofit.sourceforge.net/license.txt
[#1] INFO:Minization -- RooMinimizer::optimizeConst: activating const optimization
**********
** 1 **SET PRINT 1
**********
**********
** 2 **SET NOGRAD
**********
PARAMETER DEFINITIONS:
NO. NAME VALUE STEP SIZE LIMITS
1 mean 0.00000e+00 2.00000e+00 -1.00000e+01 1.00000e+01
2 sigma 3.00000e+00 9.90000e-01 1.00000e-01 1.00000e+01
**********
** 3 **SET ERR 0.5
**********
**********
** 4 **SET PRINT 1
**********
**********
** 5 **SET STR 1
**********
NOW USING STRATEGY 1: TRY TO BALANCE SPEED AGAINST RELIABILITY
**********
** 6 **MIGRAD 1000 1
**********
FIRST CALL TO USER FUNCTION AT NEW START POINT, WITH IFLAG=4.
START MIGRAD MINIMIZATION. STRATEGY 1. CONVERGENCE WHEN EDM .LT. 1.00e-03
FCN=249.349 FROM MIGRAD STATUS=INITIATE 8 CALLS 9 TOTAL
EDM= unknown STRATEGY= 1 NO ERROR MATRIX
EXT PARAMETER CURRENT GUESS STEP FIRST
NO. NAME VALUE ERROR SIZE DERIVATIVE
1 mean 0.00000e+00 2.00000e+00 2.01358e-01 1.15556e+01
2 sigma 3.00000e+00 9.90000e-01 2.22742e-01 5.42294e+00
ERR DEF= 0.5
MIGRAD MINIMIZATION HAS CONVERGED.
MIGRAD WILL VERIFY CONVERGENCE AND ERROR MATRIX.
COVARIANCE MATRIX CALCULATED SUCCESSFULLY
FCN=249.251 FROM MIGRAD STATUS=CONVERGED 23 CALLS 24 TOTAL
EDM=1.58964e-05 STRATEGY= 1 ERROR MATRIX ACCURATE
EXT PARAMETER STEP FIRST
NO. NAME VALUE ERROR SIZE DERIVATIVE
1 mean -1.05079e-01 2.95122e-01 3.29083e-04 -2.34747e-02
2 sigma 2.93926e+00 2.13363e-01 5.44955e-04 -8.23858e-02
ERR DEF= 0.5
EXTERNAL ERROR MATRIX. NDIM= 25 NPAR= 2 ERR DEF=0.5
8.712e-02 -9.823e-05
-9.823e-05 4.556e-02
PARAMETER CORRELATION COEFFICIENTS
NO. GLOBAL 1 2
1 0.00156 1.000 -0.002
2 0.00156 -0.002 1.000
**********
** 7 **SET ERR 0.5
**********
**********
** 8 **SET PRINT 1
**********
**********
** 9 **HESSE 1000
**********
COVARIANCE MATRIX CALCULATED SUCCESSFULLY
FCN=249.251 FROM HESSE STATUS=OK 10 CALLS 34 TOTAL
EDM=1.58906e-05 STRATEGY= 1 ERROR MATRIX ACCURATE
EXT PARAMETER INTERNAL INTERNAL
NO. NAME VALUE ERROR STEP SIZE VALUE
1 mean -1.05079e-01 2.95122e-01 6.58167e-05 -1.05081e-02
2 sigma 2.93926e+00 2.13363e-01 1.08991e-04 -4.40523e-01
ERR DEF= 0.5
EXTERNAL ERROR MATRIX. NDIM= 25 NPAR= 2 ERR DEF=0.5
8.712e-02 -1.406e-04
-1.406e-04 4.556e-02
PARAMETER CORRELATION COEFFICIENTS
NO. GLOBAL 1 2
1 0.00223 1.000 -0.002
2 0.00223 -0.002 1.000
[#1] INFO:Minization -- RooMinimizer::optimizeConst: deactivating const optimization
[#1] INFO:Eval -- RooTreeDataStore::loadValues(ds) Ignored 36 out of range events
-----------------------
Reading data from ASCII
[#1] INFO:DataHandling -- RooDataSet::read: reading file /tmp/rf102_testData.txt
[#0] ERROR:DataHandling -- RooDataSet::read(static): read error at line 65
[#1] INFO:DataHandling -- RooDataSet::read: read 64 events (ignored 0 out of range events)
DataStore dataset (/tmp/rf102_testData.txt)
Contains 64 entries
Observables:
1) x = 0.0174204 L(-10 - 10) "x"
2) y = 9.46654 L(-10 - 10) "y"
3) blindState = Normal(idx = 0)
"Blinding State"
Original data, line 20:
1) RooRealVar:: x = -0.79919
2) RooRealVar:: y = 0.0106407
Read-back data, line 20:
1) RooRealVar:: x = -0.79919
2) RooRealVar:: y = 0.0106407
3) RooCategory:: blindState = Normal(idx = 0)
RooDataSet::ds[x,y] = 64 entries
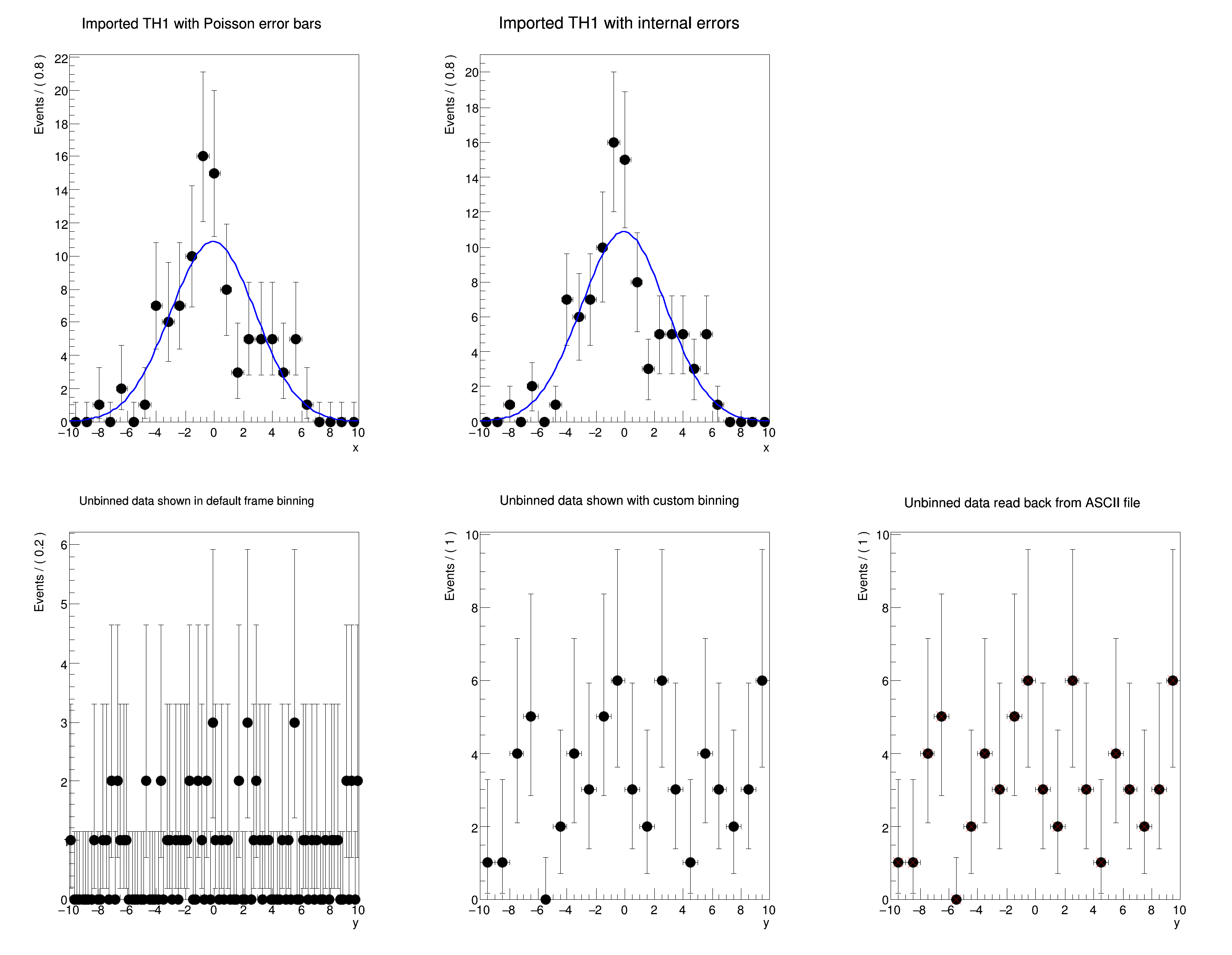
void rf102_dataimport()
{
RooPlot *frame =
x.frame(
Title(
"Imported TH1 with Poisson error bars"));
dh.plotOn(frame);
RooPlot *frame2 =
x.frame(
Title(
"Imported TH1 with internal errors"));
{
std::ofstream outstream("/tmp/rf102_testData.txt");
ds.write(outstream);
outstream.close();
}
std::cout << "\n-----------------------\nReading data from ASCII\n";
"D");
dataReadBack->
Print(
"V");
std::cout << "\nOriginal data, line 20:\n";
ds.get(20)->Print("V");
std::cout << "\nRead-back data, line 20:\n";
ds.Print();
RooPlot *frame3 =
y.frame(
Title(
"Unbinned data shown in default frame binning"));
ds.plotOn(frame3);
RooPlot *frame4 =
y.frame(
Title(
"Unbinned data shown with custom binning"));
RooPlot *frame5 =
y.frame(
Title(
"Unbinned data read back from ASCII file"));
TCanvas *
c =
new TCanvas(
"rf102_dataimport",
"rf102_dataimport", 1000, 800);
gPad->SetLeftMargin(0.15);
gPad->SetLeftMargin(0.15);
gPad->SetLeftMargin(0.15);
gPad->SetLeftMargin(0.15);
gPad->SetLeftMargin(0.15);
}
{
TH1D *hh =
new TH1D(
"hh",
"hh", 25, -10, 10);
for (int i = 0; i < 100; i++) {
}
return hh;
}
{
tree->Branch(
"x", px,
"x/D");
tree->Branch(
"y", py,
"y/D");
for (int i = 0; i < 100; i++) {
}
}
R__EXTERN TRandom * gRandom
virtual void Print(Option_t *options=0) const
This method must be overridden when a class wants to print itself.
virtual void Print(Option_t *options=0) const
Print TNamed name and title.
virtual RooPlot * plotOn(RooPlot *frame, const RooCmdArg &arg1=RooCmdArg::none(), const RooCmdArg &arg2=RooCmdArg::none(), const RooCmdArg &arg3=RooCmdArg::none(), const RooCmdArg &arg4=RooCmdArg::none(), const RooCmdArg &arg5=RooCmdArg::none(), const RooCmdArg &arg6=RooCmdArg::none(), const RooCmdArg &arg7=RooCmdArg::none(), const RooCmdArg &arg8=RooCmdArg::none()) const
Calls RooPlot* plotOn(RooPlot* frame, const RooLinkedList& cmdList) const ;.
RooArgList is a container object that can hold multiple RooAbsArg objects.
RooArgSet is a container object that can hold multiple RooAbsArg objects.
The RooDataHist is a container class to hold N-dimensional binned data.
RooDataSet is a container class to hold unbinned data.
virtual const RooArgSet * get(Int_t index) const
Return RooArgSet with coordinates of event 'index'.
static RooDataSet * read(const char *filename, const RooArgList &variables, const char *opts="", const char *commonPath="", const char *indexCatName=0)
Read given list of ascii files, and construct a data set, using the given ArgList as structure defini...
A RooPlot is a plot frame and a container for graphics objects within that frame.
virtual void Draw(Option_t *options=0)
Draw this plot and all of the elements it contains.
RooRealVar represents a fundamental (non-derived) real valued object.
virtual void SetTitleOffset(Float_t offset=1)
Set distance between the axis and the axis title Offset is a correction factor with respect to the "s...
1-D histogram with a double per channel (see TH1 documentation)}
virtual Int_t Fill(Double_t x)
Increment bin with abscissa X by 1.
virtual Double_t Gaus(Double_t mean=0, Double_t sigma=1)
Samples a random number from the standard Normal (Gaussian) Distribution with the given mean and sigm...
virtual Double_t Uniform(Double_t x1=1)
Returns a uniform deviate on the interval (0, x1).
A TTree represents a columnar dataset.
Template specialisation used in RooAbsArg:
RooCmdArg MarkerColor(Color_t color)
RooCmdArg Binning(const RooAbsBinning &binning)
RooCmdArg MarkerStyle(Style_t style)
RooCmdArg Import(const char *state, TH1 &histo)
RooCmdArg DataError(Int_t)
static constexpr double gauss



 Basic functionality: importing data from ROOT TTrees and THx histograms
Basic functionality: importing data from ROOT TTrees and THx histograms