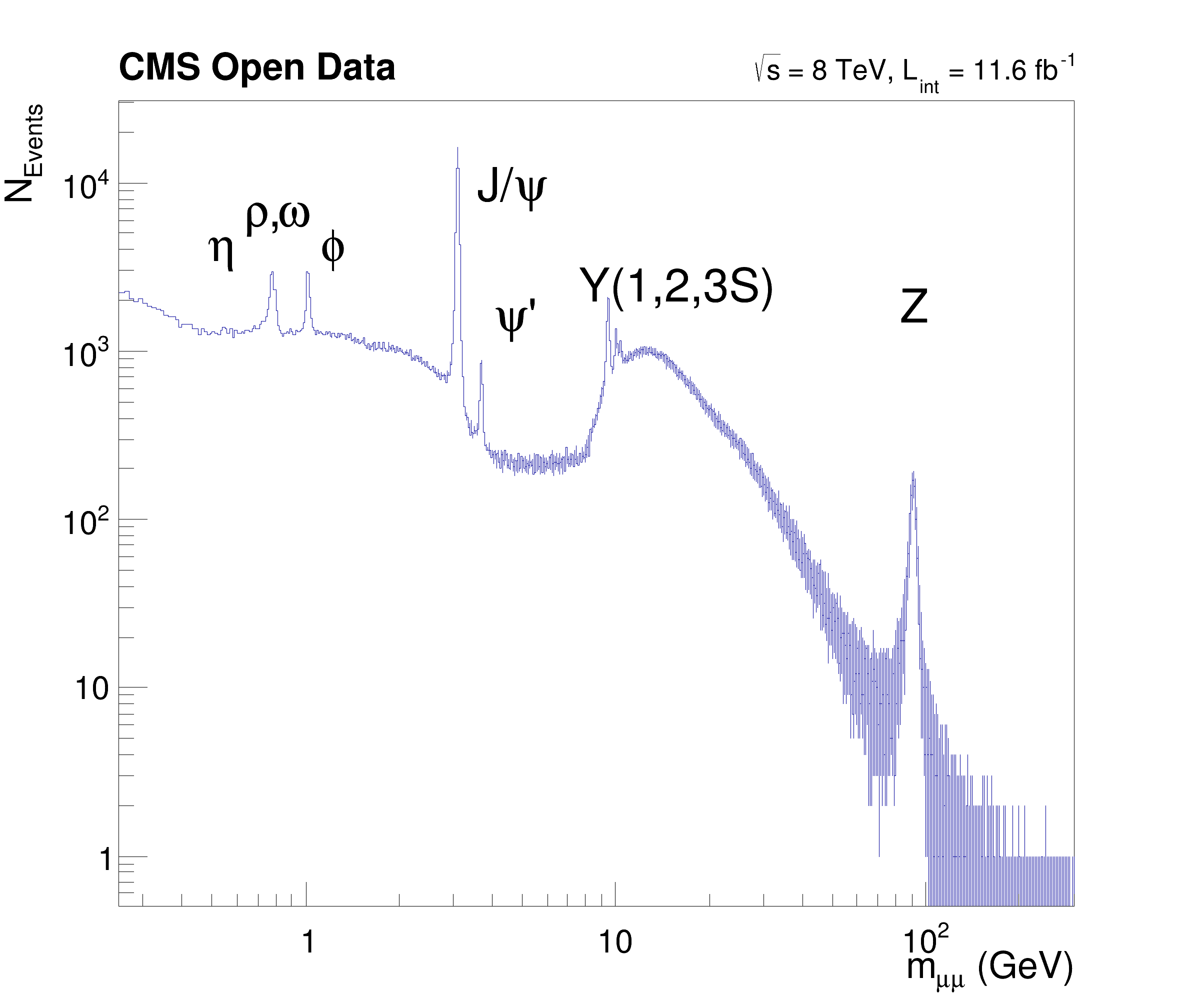
This tutorial illustrates data conversion and data processing with RNTuple and RDataFrame. In contrast to the LHCb open data tutorial, the data model in this tutorial is not tabular but entries have variable lengths vectors Based on RDataFrame's df102_NanoAODDimuonAnalysis.C
#include <cassert>
#include <cmath>
#include <iostream>
#include <memory>
#include <string>
#include <vector>
#include <utility>
constexpr char const* kTreeFileName = "http://root.cern.ch/files/NanoAOD_DoubleMuon_CMS2011OpenData.root";
constexpr char const* kNTupleFileName = "ntpl004_dimuon.root";
using ColNames_t = std::vector<std::string>;
template <typename... ColumnTypes_t>
class RNTupleHelper : public ROOT::Detail::RDF::RActionImpl<RNTupleHelper<ColumnTypes_t...>> {
public:
using Result_t = RNTupleWriter;
private:
using ColumnValues_t = std::tuple<std::shared_ptr<ColumnTypes_t>...>;
std::string fNTupleName;
std::string fRootFile;
ColNames_t fColNames;
ColumnValues_t fColumnValues;
static constexpr const auto fNColumns = std::tuple_size<ColumnValues_t>::value;
std::shared_ptr<RNTupleWriter> fNTuple;
int fCounter;
template<std::size_t...
S>
void InitializeImpl(std::index_sequence<S...>) {
std::initializer_list<int> expander{
(std::get<S>(fColumnValues) = eventModel->MakeField<ColumnTypes_t>(fColNames[
S]), 0)...};
fNTuple = std::move(RNTupleWriter::Recreate(std::move(eventModel), fNTupleName, fRootFile));
}
template<std::size_t...
S>
void ExecImpl(std::index_sequence<S...>, ColumnTypes_t... values) {
std::initializer_list<int> expander{(*std::get<S>(fColumnValues) = values, 0)...};
}
public:
: fNTupleName(ntupleName), fRootFile(rootFile), fColNames(colNames)
{
InitializeImpl(std::make_index_sequence<fNColumns>());
}
RNTupleHelper(RNTupleHelper&&) = default;
RNTupleHelper(const RNTupleHelper&) = delete;
std::shared_ptr<RNTupleWriter> GetResultPtr() const { return fNTuple; }
{
fCounter = 0;
}
void Exec(unsigned int slot, ColumnTypes_t... values)
{
ExecImpl(std::make_index_sequence<fNColumns>(), values...);
fNTuple->Fill();
if (++fCounter % 100000 == 0)
std::cout << "Wrote " << fCounter << " entries" << std::endl;
}
void Finalize()
{
fNTuple->CommitCluster();
}
std::string GetActionName() { return "RNTuple Writer"; }
};
template <typename T>
T InvariantMassStdVector(std::vector<T>&
pt, std::vector<T>& eta, std::vector<T>& phi, std::vector<T>& mass)
{
assert(
pt.size() == 2 && eta.size() == 2 && phi.size() == 2 && mass.size() == 2);
}
void Convert() {
std::string typeList = "<";
std::string columnList = "{";
auto columnNames = df.GetColumnNames();
for (
auto name : columnNames) {
auto typeName = df.GetColumnType(
name);
if (typeName == "ULong64_t") continue;
columnList +=
"\"" +
name +
"\",";
typeList += typeName + ",";
}
*columnList.rbegin() = '}';
*typeList.rbegin() = '>';
std::string code = "{";
code += "auto df = std::make_unique<ROOT::RDataFrame>(\"Events\", \"" + std::string(kTreeFileName)
+ "\")->Range(0, 4000000);";
code += "ColNames_t colNames = " + columnList + ";";
code += "RNTupleHelper" + typeList + " helper{\"Events\", \"" + std::string(kNTupleFileName) + "\", colNames};";
code += "*df.Book" + typeList + "(std::move(helper), colNames);";
code += "}";
}
void ntpl004_dimuon() {
Convert();
auto df_2mu = df.Filter("nMuon == 2", "Events with exactly two muons");
auto df_os = df_2mu.Filter("Muon_charge[0] != Muon_charge[1]", "Muons with opposite charge");
auto df_mass = df_os.Define("Dimuon_mass", InvariantMassStdVector<float>, {"Muon_pt", "Muon_eta", "Muon_phi", "Muon_mass"});
auto df_size = df_os.Define("eta_size", "Muon_mass.size()");
auto h = df_mass.Histo1D({
"Dimuon_mass",
"Dimuon_mass", 30000, 0.25, 300},
"Dimuon_mass");
auto report = df_mass.Report();
auto c =
new TCanvas(
"c",
"", 800, 700);
c->SetLogx();
c->SetLogy();
h->GetXaxis()->SetTitle(
"m_{#mu#mu} (GeV)");
h->GetXaxis()->SetTitleSize(0.04);
h->GetYaxis()->SetTitle(
"N_{Events}");
h->GetYaxis()->SetTitleSize(0.04);
label.
DrawLatex(0.205, 0.775,
"#rho,#omega");
label.
SetTextSize(0.030); label.
DrawLatex(0.630, 0.920,
"#sqrt{s} = 8 TeV, L_{int} = 11.6 fb^{-1}");
}
R__EXTERN TStyle * gStyle
R__EXTERN TSystem * gSystem
static std::unique_ptr< RNTupleModel > Create()
An RNTuple that is used to read data from storage.
An RNTuple that gets filled with entries (data) and writes them to storage.
ROOT's RDataFrame offers a high level interface for analyses of data stored in TTrees,...
A "std::vector"-like collection of values implementing handy operation to analyse them.
virtual void SetTextFont(Font_t tfont=62)
Set the text font.
virtual void SetTextSize(Float_t tsize=1)
Set the text size.
To draw Mathematical Formula.
TLatex * DrawLatex(Double_t x, Double_t y, const char *text)
Make a copy of this object with the new parameters And copy object attributes.
void SetOptStat(Int_t stat=1)
The type of information printed in the histogram statistics box can be selected via the parameter mod...
virtual Bool_t AccessPathName(const char *path, EAccessMode mode=kFileExists)
Returns FALSE if one can access a file using the specified access mode.
virtual void Print(Option_t *option="") const
Dump this text with its attributes.
virtual void SetNDC(Bool_t isNDC=kTRUE)
Set NDC mode on if isNDC = kTRUE, off otherwise.
A simple, robust and fast interface to read values from ROOT columnar datasets such as TTree,...
basic_string_view< char > string_view
RDataFrame MakeNTupleDataFrame(std::string_view ntupleName, std::string_view fileName)
Vector1::Scalar InvariantMass(const Vector1 &v1, const Vector2 &v2)
return the invariant mass of two LorentzVector The only requirement on the LorentzVector is that they...
RooArgSet S(const RooAbsArg &v1)
void Initialize(Bool_t useTMVAStyle=kTRUE)



 Convert CMS open data from a TTree to RNTuple.
Convert CMS open data from a TTree to RNTuple.