
 Standard demo of the Profile Likelihood calculator StandardProfileLikelihoodDemo
Standard demo of the Profile Likelihood calculator StandardProfileLikelihoodDemo
This is a standard demo that can be used with any ROOT file prepared in the standard way. You specify:
- name for input ROOT file
- name of workspace inside ROOT file that holds model and data
- name of ModelConfig that specifies details for calculator tools
- name of dataset
With default parameters the macro will attempt to run the standard hist2workspace example and read the ROOT file that it produces.
The actual heart of the demo is only about 10 lines long.
The ProfileLikelihoodCalculator is based on Wilks's theorem and the asymptotic properties of the profile likelihood ratio (eg. that it is chi-square distributed for the true value).
␛[1mRooFit v3.60 -- Developed by Wouter Verkerke and David Kirkby␛[0m
Copyright (C) 2000-2013 NIKHEF, University of California & Stanford University
All rights reserved, please read http://roofit.sourceforge.net/license.txt
[#1] INFO:Minization -- p.d.f. provides expected number of events, including extended term in likelihood.
[#1] INFO:Minization -- createNLL: caching constraint set under name CONSTR_OF_PDF_simPdf_FOR_OBS_channelCat:obs_x_channel1 with 4 entries
[#1] INFO:Minization -- Including the following contraint terms in minimization: (alpha_syst2Constraint,alpha_syst3Constraint,gamma_stat_channel1_bin_0_constraint,gamma_stat_channel1_bin_1_constraint)
[#1] INFO:Minization -- The following global observables have been defined: (nom_alpha_syst2,nom_alpha_syst3,nom_gamma_stat_channel1_bin_0,nom_gamma_stat_channel1_bin_1)
[#0] PROGRESS:Minization -- ProfileLikelihoodCalcultor::DoGLobalFit - find MLE
[#0] PROGRESS:Minization -- ProfileLikelihoodCalcultor::DoMinimizeNLL - using Minuit / Migrad with strategy 1
[#1] INFO:Fitting -- RooAddition::defaultErrorLevel(nll_simPdf_obsData_with_constr) Summation contains a RooNLLVar, using its error level
[#1] INFO:Minization -- RooMinimizer::optimizeConst: activating const optimization
RooAbsTestStatistic::initSimMode: creating slave calculator #0 for state channel1 (2 dataset entries)
[#1] INFO:NumericIntegration -- RooRealIntegral::init(channel1_model_Int[obs_x_channel1]) using numeric integrator RooBinIntegrator to calculate Int(obs_x_channel1)
[#1] INFO:Fitting -- RooAbsTestStatistic::initSimMode: created 1 slave calculators.
[#1] INFO:NumericIntegration -- RooRealIntegral::init(channel1_model_Int[obs_x_channel1]) using numeric integrator RooBinIntegrator to calculate Int(obs_x_channel1)
[#1] INFO:Minization -- The following expressions have been identified as constant and will be precalculated and cached: (signal_channel1_nominal,background1_channel1_nominal,background2_channel1_nominal)
[#1] INFO:Minization -- The following expressions will be evaluated in cache-and-track mode: (mc_stat_channel1)
[#1] INFO:Minization --
RooFitResult: minimized FCN value: -1033.78, estimated distance to minimum: 2.39561e-08
covariance matrix quality: Full, accurate covariance matrix
Status : MINIMIZE=0
Floating Parameter FinalValue +/- Error
-------------------- --------------------------
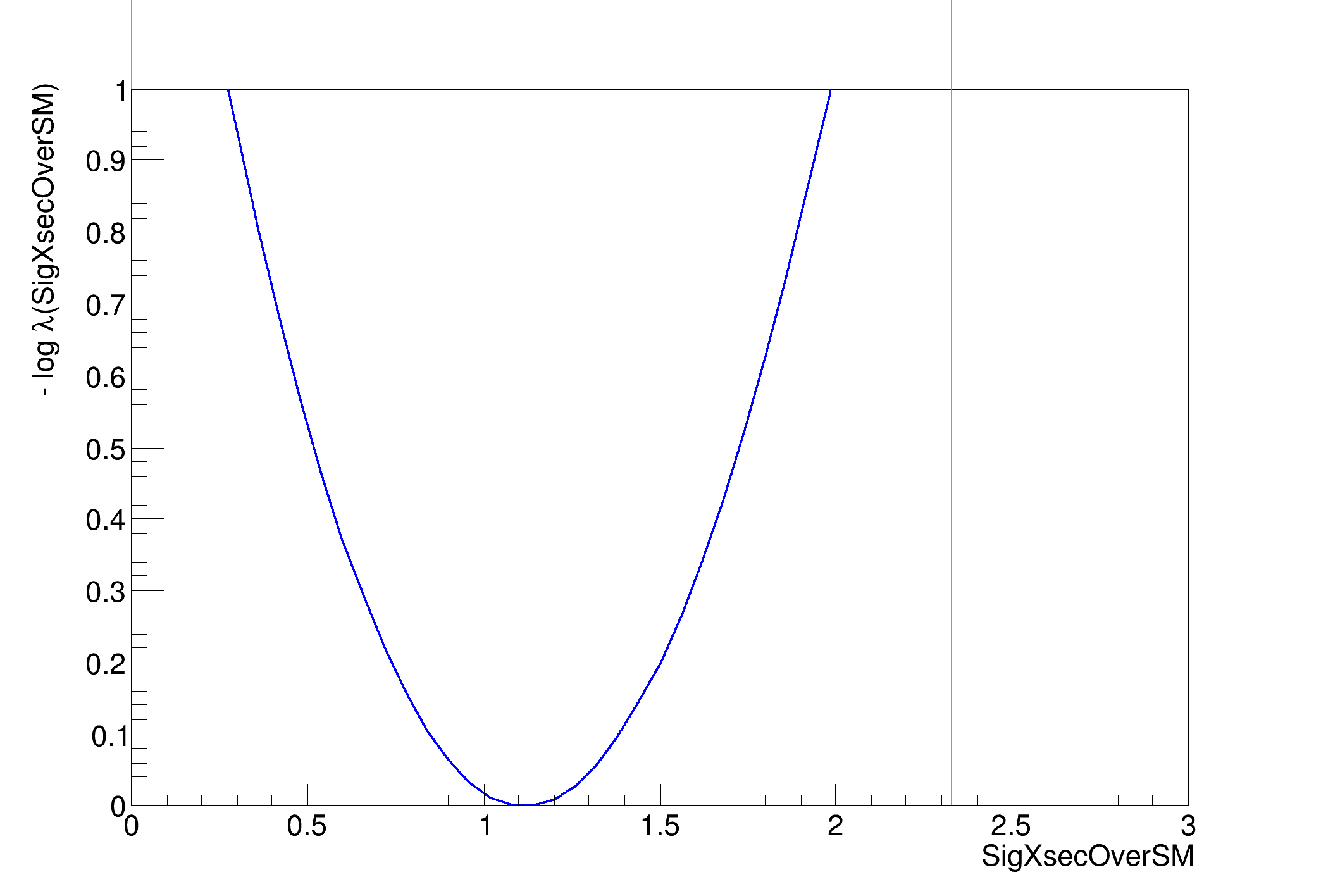
SigXsecOverSM 1.1153e+00 +/- 5.86e-01
alpha_syst2 -8.9017e-03 +/- 9.82e-01
alpha_syst3 1.7918e-02 +/- 9.48e-01
gamma_stat_channel1_bin_0 9.9955e-01 +/- 4.93e-02
gamma_stat_channel1_bin_1 1.0036e+00 +/- 8.01e-02
>>>> RESULT : 95% interval on SigXsecOverSM is : [Warning: lower value for SigXsecOverSM is at limit 0
0, 2.32744]
making a plot of the profile likelihood function ....(if it is taking a lot of time use less points or the TF1 drawing option)
[#1] INFO:Minization -- RooProfileLL::evaluate(nll_simPdf_obsData_with_constr_Profile[SigXsecOverSM]) Creating instance of MINUIT
[#1] INFO:Fitting -- RooAddition::defaultErrorLevel(nll_simPdf_obsData_with_constr) Summation contains a RooNLLVar, using its error level
[#1] INFO:Minization -- RooProfileLL::evaluate(nll_simPdf_obsData_with_constr_Profile[SigXsecOverSM]) determining minimum likelihood for current configurations w.r.t all observable
[#1] INFO:Minization -- RooProfileLL::evaluate(nll_simPdf_obsData_with_constr_Profile[SigXsecOverSM]) minimum found at (SigXsecOverSM=1.11511)
.
[#1] INFO:Minization -- RooProfileLL::evaluate(nll_simPdf_obsData_with_constr_Profile[SigXsecOverSM]) Creating instance of MINUIT
[#1] INFO:Fitting -- RooAddition::defaultErrorLevel(nll_simPdf_obsData_with_constr) Summation contains a RooNLLVar, using its error level
[#1] INFO:Minization -- RooProfileLL::evaluate(nll_simPdf_obsData_with_constr_Profile[SigXsecOverSM]) determining minimum likelihood for current configurations w.r.t all observable
RooAbsTestStatistic::initSimMode: creating slave calculator #0 for state channel1 (2 dataset entries)
[#1] INFO:NumericIntegration -- RooRealIntegral::init(channel1_model_Int[obs_x_channel1]) using numeric integrator RooBinIntegrator to calculate Int(obs_x_channel1)
[#1] INFO:Fitting -- RooAbsTestStatistic::initSimMode: created 1 slave calculators.
[#1] INFO:NumericIntegration -- RooRealIntegral::init(channel1_model_Int[obs_x_channel1]) using numeric integrator RooBinIntegrator to calculate Int(obs_x_channel1)
[#0] ERROR:InputArguments -- RooArgSet::checkForDup: ERROR argument with name SigXsecOverSM is already in this set
[#1] INFO:Minization -- RooProfileLL::evaluate(nll_simPdf_obsData_with_constr_Profile[SigXsecOverSM]) minimum found at (SigXsecOverSM=1.11509)
......................................................................................................
struct ProfileLikelihoodOptions {
double confLevel = 0.95;
int nScanPoints = 50;
bool plotAsTF1 = false;
double poiMinPlot = 1;
double poiMaxPlot = 0;
bool doHypoTest = false;
double nullValue = 0;
};
ProfileLikelihoodOptions optPL;
void StandardProfileLikelihoodDemo(const char *infile = "", const char *workspaceName = "combined",
const char *modelConfigName = "ModelConfig", const char *dataName = "obsData")
{
double confLevel = optPL.confLevel;
double nScanPoints = optPL.nScanPoints;
bool plotAsTF1 = optPL.plotAsTF1;
double poiXMin = optPL.poiMinPlot;
double poiXMax = optPL.poiMaxPlot;
bool doHypoTest = optPL.doHypoTest;
double nullParamValue = optPL.nullValue;
const char *filename = "";
if (!strcmp(infile, "")) {
filename = "results/example_combined_GaussExample_model.root";
if (!fileExist) {
#ifdef _WIN32
cout << "HistFactory file cannot be generated on Windows - exit" << endl;
return;
#endif
cout << "will run standard hist2workspace example" << endl;
gROOT->ProcessLine(
".! prepareHistFactory .");
gROOT->ProcessLine(
".! hist2workspace config/example.xml");
cout << "\n\n---------------------" << endl;
cout << "Done creating example input" << endl;
cout << "---------------------\n\n" << endl;
}
} else
filename = infile;
cout << "StandardRooStatsDemoMacro: Input file " << filename << " is not found" << endl;
return;
}
if (!w) {
cout << "workspace not found" << endl;
return;
}
ModelConfig *mc = (ModelConfig *)w->
obj(modelConfigName);
if (!data || !mc) {
cout << "data or ModelConfig was not found" << endl;
return;
}
ProfileLikelihoodCalculator pl(*data, *mc);
pl.SetConfidenceLevel(confLevel);
LikelihoodInterval *interval = pl.GetInterval();
cout <<
"\n>>>> RESULT : " << confLevel * 100 <<
"% interval on " << firstPOI->
GetName() <<
" is : [" << interval->LowerLimit(*firstPOI) << ", " << interval->UpperLimit(*firstPOI) << "]\n " << endl;
cout << "making a plot of the profile likelihood function ....(if it is taking a lot of time use less points or the "
"TF1 drawing option)\n";
LikelihoodIntervalPlot plot(interval);
plot.SetNPoints(nScanPoints);
if (poiXMin < poiXMax)
plot.SetRange(poiXMin, poiXMax);
if (plotAsTF1)
plot.Draw(opt);
if (doHypoTest) {
nullparams.addClone(*firstPOI);
nullparams.setRealValue(firstPOI->
GetName(), nullParamValue);
pl.SetNullParameters(nullparams);
std::cout <<
"Perform Test of Hypothesis : null Hypothesis is " << firstPOI->
GetName() << nullParamValue
<< std::endl;
auto result = pl.GetHypoTest();
std::cout << "\n>>>> Hypotheis Test Result \n";
result->Print();
}
}
R__EXTERN TSystem * gSystem
RooAbsData is the common abstract base class for binned and unbinned datasets.
RooArgSet is a container object that can hold multiple RooAbsArg objects.
RooRealVar represents a fundamental (non-derived) real valued object.
The RooWorkspace is a persistable container for RooFit projects.
RooAbsData * data(const char *name) const
Retrieve dataset (binned or unbinned) with given name. A null pointer is returned if not found.
void Print(Option_t *opts=0) const
Print contents of the workspace.
TObject * obj(const char *name) const
Return any type of object (RooAbsArg, RooAbsData or generic object) with given name)
A ROOT file is a suite of consecutive data records (TKey instances) with a well defined format.
static TFile * Open(const char *name, Option_t *option="", const char *ftitle="", Int_t compress=ROOT::RCompressionSetting::EDefaults::kUseGeneralPurpose, Int_t netopt=0)
Create / open a file.
virtual const char * GetName() const
Returns name of object.
virtual Bool_t AccessPathName(const char *path, EAccessMode mode=kFileExists)
Returns FALSE if one can access a file using the specified access mode.
Template specialisation used in RooAbsArg:
Namespace for the RooStats classes.
- Author
- Kyle Cranmer
Definition in file StandardProfileLikelihoodDemo.C.



 Standard demo of the Profile Likelihood calculator StandardProfileLikelihoodDemo
Standard demo of the Profile Likelihood calculator StandardProfileLikelihoodDemo 