Processing /mnt/build/workspace/root-makedoc-v614/rootspi/rdoc/src/v6-14-00-patches/tutorials/roofit/rf402_datahandling.C...
�[1mRooFit v3.60 -- Developed by Wouter Verkerke and David Kirkby�[0m
Copyright (C) 2000-2013 NIKHEF, University of California & Stanford University
All rights reserved, please read http://roofit.sourceforge.net/license.txt
DataStore d (d)
Contains 1000 entries
Observables:
1) x = 9 L(-10 - 10) "x"
2) y = 31.607 L(0 - 40) "y"
3) c = Plus(idx = 1)
"c"
1) 0x260f820 RooRealVar:: x = 9 L(-10 - 10) "x"
2) 0x23ac470 RooRealVar:: y = 31.607 L(0 - 40) "y"
3) 0x2679380 RooCategory:: c = Plus(idx = 1)
"c"
1) 0x260f820 RooRealVar:: x = 8 L(-10 - 10) "x"
2) 0x23ac470 RooRealVar:: y = 30 L(0 - 40) "y"
3) 0x2679380 RooCategory:: c = Minus(idx = -1)
"c"
>> d1 has only columns x,c
DataStore d (d)
Contains 1000 entries
Observables:
1) x = 9 L(-10 - 10) "x"
2) c = Plus(idx = 1)
"c"
>> d2 has only column y
DataStore d (d)
Contains 1000 entries
Observables:
1) y = 31.607 L(0 - 40) "y"
>> d3 has only the points with y>5.17
DataStore d (d)
Contains 973 entries
Observables:
1) x = 9 L(-10 - 10) "x"
2) y = 31.607 L(0 - 40) "y"
3) c = Plus(idx = 1)
"c"
>> d4 has only columns x,c for data points with y>5.17
DataStore d (d)
Contains 973 entries
Observables:
1) x = 9 L(-10 - 10) "x"
2) c = Plus(idx = 1)
"c"
>> merge d2(y) with d1(x,c) to form d1(x,c,y)
DataStore d (d)
Contains 1000 entries
Observables:
1) x = 9 L(-10 - 10) "x"
2) c = Plus(idx = 1)
"c"
3) y = 31.607 L(0 - 40) "y"
>> append data points of d3 to d1
DataStore d (d)
Contains 1973 entries
Observables:
1) x = 9 L(-10 - 10) "x"
2) c = Plus(idx = 1)
"c"
3) y = 31.607 L(0 - 40) "y"
>> construct dh (binned) from d(unbinned) but only take the x and y dimensions,
>> the category 'c' will be projected in the filling process
DataStore dh (binned version of d)
Contains 100 entries
Observables:
1) x = 9 L(-10 - 10) B(10) "x"
2) y = 31.607 L(0 - 40) B(10) "y"
Binned Dataset dh (binned version of d)
Contains 100 bins with a total weight of 1000
Observables: 1) x = 9 L(-10 - 10) B(10) "x"
2) y = 31.607 L(0 - 40) B(10) "y"
>> number of bins in dh : 100
>> sum of weights in dh : 1000
>> integral over histogram: 8000
>> retrieving the properties of the bin enclosing coordinate (x,y) = (0.3,20.5)
bin center:
1) 0x2385430 RooRealVar:: x = 1 L(-10 - 10) B(10) "x"
2) 0x23f4960 RooRealVar:: y = 22 L(0 - 40) B(10) "y"
weight = 76
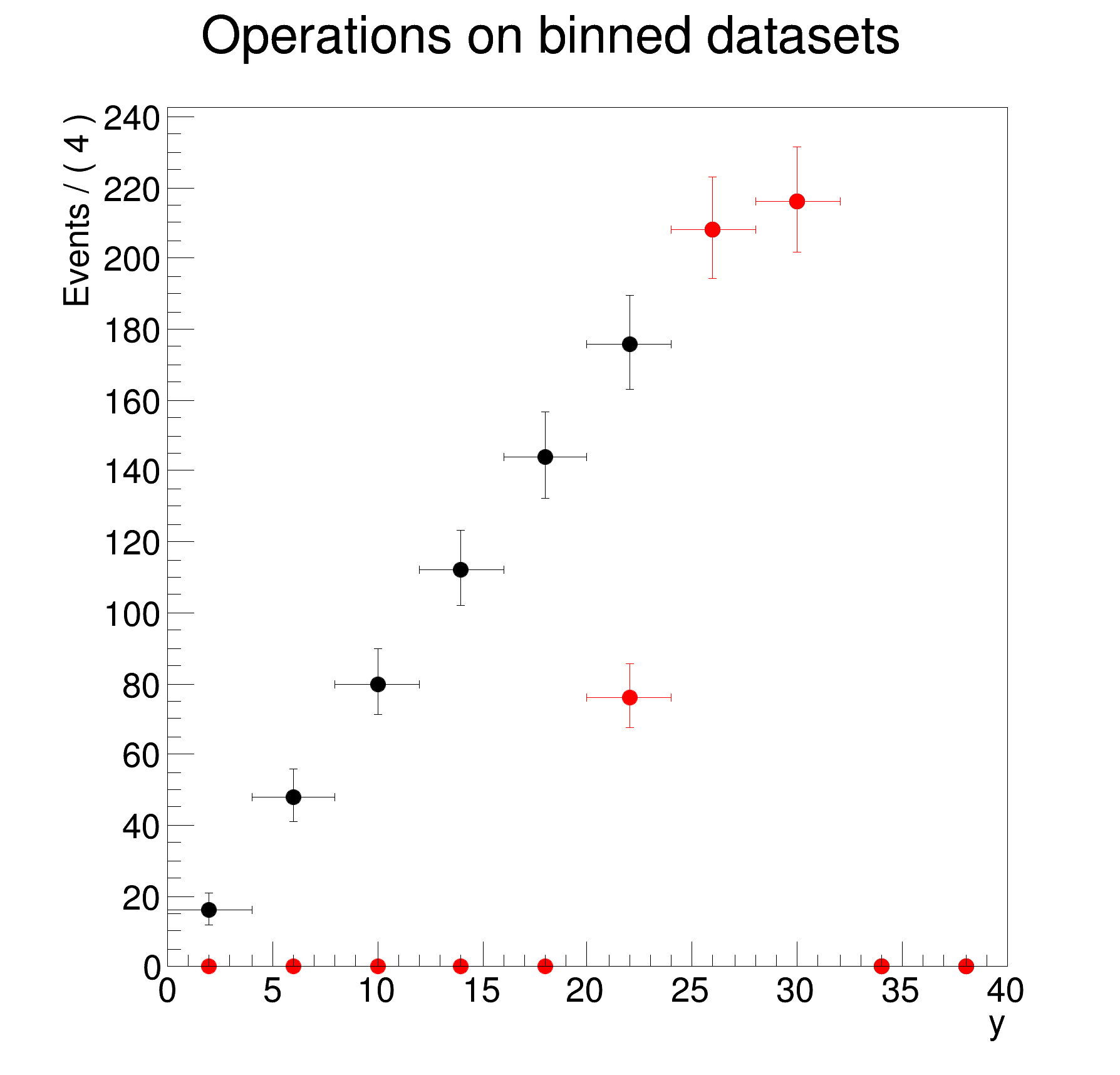
>> Creating 1-dimensional projection on y of dh for bins with x>0
DataStore dh (binned version of d)
Contains 10 entries
Observables:
1) y = 38 L(0 - 40) B(10) "y"
Binned Dataset dh (binned version of d)
Contains 10 bins with a total weight of 500
Observables: 1) y = 38 L(0 - 40) B(10) "y"
[#1] INFO:Plotting -- RooPlot::updateFitRangeNorm: New event count of 500 will supercede previous event count of 1000 for normalization of PDF projections
>> Persisting d via ROOT I/O
TFile** rf402_datahandling.root
TFile* rf402_datahandling.root
KEY: RooDataSet d;1 d
KEY: TProcessID ProcessID0;1 3920539e-de7d-11e8-90cd-e2c88e80beef
void rf402_datahandling()
{
c.defineType("Plus",+1) ;
c.defineType("Minus",-1) ;
for (i=0 ; i<1000 ; i++) {
x = i/50 - 10 ;
c.setLabel((i%2)?"Plus":"Minus") ;
}
cout << endl ;
cout << endl ;
cout << endl ;
cout << endl << ">> d1 has only columns x,c" << endl ;
cout << endl << ">> d2 has only column y" << endl ;
cout << endl << ">> d3 has only the points with y>5.17" << endl ;
cout << endl << ">> d4 has only columns x,c for data points with y>5.17" << endl ;
cout << endl << ">> merge d2(y) with d1(x,c) to form d1(x,c,y)" << endl ;
cout << endl << ">> append data points of d3 to d1" << endl ;
cout << ">> construct dh (binned) from d(unbinned) but only take the x and y dimensions," << endl
<< ">> the category 'c' will be projected in the filling process" << endl ;
x.setBins(10) ;
y.setBins(10) ;
cout <<
">> number of bins in dh : " << dh.
numEntries() << endl ;
cout <<
">> sum of weights in dh : " << dh.
sum(
kFALSE) << endl ;
cout <<
">> integral over histogram: " << dh.
sum(
kTRUE) << endl ;
x = 0.3 ; y = 20.5 ;
cout << ">> retrieving the properties of the bin enclosing coordinate (x,y) = (0.3,20.5) " << endl ;
cout << " bin center:" << endl ;
cout <<
" weight = " << dh.
weight() << endl ;
cout << ">> Creating 1-dimensional projection on y of dh for bins with x>0" << endl ;
cout << endl << ">> Persisting d via ROOT I/O" << endl ;
TFile
f(
"rf402_datahandling.root",
"RECREATE") ;
new TCanvas(
"rf402_datahandling",
"rf402_datahandling",600,600) ;
}



 'DATA AND CATEGORIES' RooFit tutorial macro #402
'DATA AND CATEGORIES' RooFit tutorial macro #402