[#0] PROGRESS:InputArguments -- initializing physics inputs from file /github/home/ROOT-CI/build/tutorials/roofit/roofit/input_histos_rf_lagrangianmorph.root with object name(s) 'pTV'
[#0] PROGRESS:Caching -- creating cache from getCache function for 0x560cc70268f0
[#0] PROGRESS:Caching -- current storage has size 10
[#0] PROGRESS:ObjectHandling -- observable: pTV
[#0] PROGRESS:ObjectHandling -- binWidth: binWidth_pTV
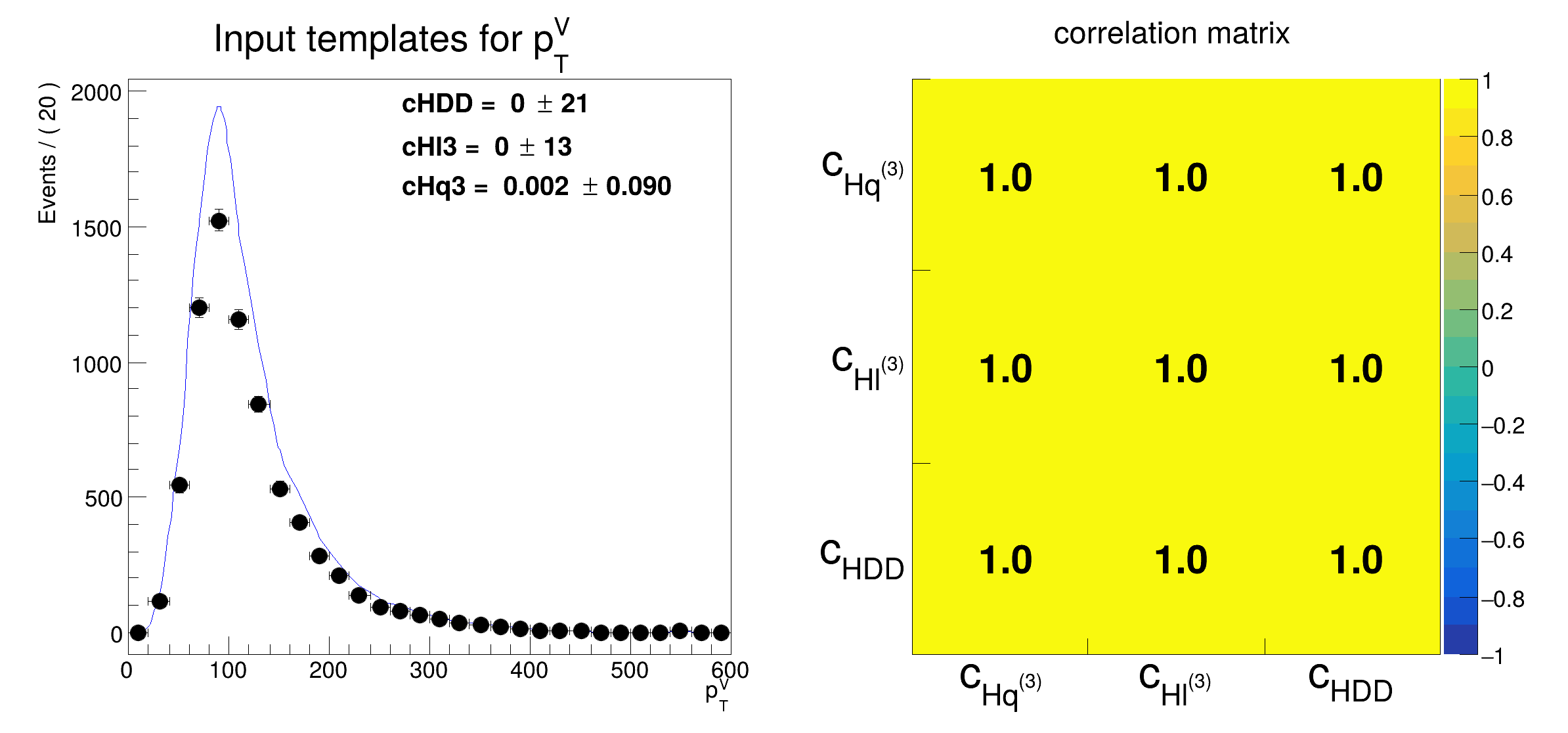
[#1] INFO:DataHandling -- RooDataHist::adjustBinning(pseudo_dh): fit range of variable pTV expanded to nearest bin boundaries: [10,600] --> [0,600]
[#0] PROGRESS:Caching -- creating cache from getCache function for 0x560cc80cedb0
[#0] PROGRESS:Caching -- current storage has size 10
[#0] PROGRESS:ObjectHandling -- observable: pTV
[#0] PROGRESS:ObjectHandling -- binWidth: binWidth_pTV
[#1] INFO:Fitting -- RooAbsPdf::fitTo(wrap_pdf_over_wrap_pdf_Int[pTV]) fixing normalization set for coefficient determination to observables in data
[#1] INFO:Fitting -- using generic CPU library compiled with no vectorizations
[#1] INFO:Fitting -- Creation of NLL object took 7.73192 ms
[#1] INFO:Fitting -- RooAddition::defaultErrorLevel(nll_wrap_pdf_over_wrap_pdf_Int[pTV]_pseudo_dh) Summation contains a RooNLLVar, using its error level
[#1] INFO:Minimization -- [fitFCN] No discrete parameters, performing continuous minimization only
RooAbsMinimizerFcn: Minimized function has error status.
Returning maximum FCN so far (86348.3) to force MIGRAD to back out of this region. Error log follows.
Parameter values: cHDD=0 cHl3=0 cHq3=-0.0202918
RooFit::Detail::RooNormalizedPdf::wrap_pdf_over_wrap_pdf_Int[pTV][ numerator=wrap_pdf denominator=wrap_pdf_Int[pTV] ]
p.d.f value is less than zero, trying to recover @ numerator=wrap_pdf=0.0376648, denominator=wrap_pdf_Int[pTV]=10358.6
p.d.f value is less than zero, trying to recover @ numerator=wrap_pdf=0.0376648, denominator=wrap_pdf_Int[pTV]=10358.6
getLogVal() top-level p.d.f evaluates to NaN @ numerator=wrap_pdf=0.0376648, denominator=wrap_pdf_Int[pTV]=10358.6
getLogVal() top-level p.d.f evaluates to NaN @ numerator=wrap_pdf=0.0376648, denominator=wrap_pdf_Int[pTV]=10358.6
RooAbsMinimizerFcn: Minimized function has error status.
Returning maximum FCN so far (86348.3) to force MIGRAD to back out of this region. Error log follows.
Parameter values: cHDD=0.213672 cHl3=1.97898 cHq3=0.00773174
RooFit::Detail::RooNormalizedPdf::wrap_pdf_over_wrap_pdf_Int[pTV][ numerator=wrap_pdf denominator=wrap_pdf_Int[pTV] ]
p.d.f value is less than zero, trying to recover @ numerator=wrap_pdf=0.0882577, denominator=wrap_pdf_Int[pTV]=4536.67
p.d.f value is less than zero, trying to recover @ numerator=wrap_pdf=0.0882577, denominator=wrap_pdf_Int[pTV]=4536.67
getLogVal() top-level p.d.f evaluates to NaN @ numerator=wrap_pdf=0.0882577, denominator=wrap_pdf_Int[pTV]=4536.67
getLogVal() top-level p.d.f evaluates to NaN @ numerator=wrap_pdf=0.0882577, denominator=wrap_pdf_Int[pTV]=4536.67
RooAbsMinimizerFcn: Minimized function has error status.
Returning maximum FCN so far (86348.3) to force MIGRAD to back out of this region. Error log follows.
Parameter values: cHDD=0.0861399 cHl3=-9.50561 cHq3=0.0801661
RooFit::Detail::RooNormalizedPdf::wrap_pdf_over_wrap_pdf_Int[pTV][ numerator=wrap_pdf denominator=wrap_pdf_Int[pTV] ]
p.d.f value is less than zero, trying to recover @ numerator=wrap_pdf=-0.675078, denominator=wrap_pdf_Int[pTV]=86190.2
p.d.f value is less than zero, trying to recover @ numerator=wrap_pdf=-0.675078, denominator=wrap_pdf_Int[pTV]=86190.2
getLogVal() top-level p.d.f evaluates to NaN @ numerator=wrap_pdf=-0.675078, denominator=wrap_pdf_Int[pTV]=86190.2
getLogVal() top-level p.d.f evaluates to NaN @ numerator=wrap_pdf=-0.675078, denominator=wrap_pdf_Int[pTV]=86190.2
RooAbsMinimizerFcn: Minimized function has error status.
Returning maximum FCN so far (86348.3) to force MIGRAD to back out of this region. Error log follows.
Parameter values: cHDD=0.186765 cHl3=8.8591 cHq3=-0.971282
RooFit::Detail::RooNormalizedPdf::wrap_pdf_over_wrap_pdf_Int[pTV][ numerator=wrap_pdf denominator=wrap_pdf_Int[pTV] ]
getLogVal() top-level p.d.f evaluates to NaN @ numerator=wrap_pdf=-6.32705, denominator=wrap_pdf_Int[pTV]=46316
RooAbsMinimizerFcn: Minimized function has error status.
Returning maximum FCN so far (86348.3) to force MIGRAD to back out of this region. Error log follows.
Parameter values: cHDD=0.218731 cHl3=0.37397 cHq3=-2.08166
RooFit::Detail::RooNormalizedPdf::wrap_pdf_over_wrap_pdf_Int[pTV][ numerator=wrap_pdf denominator=wrap_pdf_Int[pTV] ]
p.d.f value is less than zero, trying to recover @ numerator=wrap_pdf=38.6705, denominator=wrap_pdf_Int[pTV]=90131.3
p.d.f value is less than zero, trying to recover @ numerator=wrap_pdf=38.6705, denominator=wrap_pdf_Int[pTV]=90131.3
getLogVal() top-level p.d.f evaluates to NaN @ numerator=wrap_pdf=38.6705, denominator=wrap_pdf_Int[pTV]=90131.3
getLogVal() top-level p.d.f evaluates to NaN @ numerator=wrap_pdf=38.6705, denominator=wrap_pdf_Int[pTV]=90131.3
RooAbsMinimizerFcn: Minimized function has error status.
Returning maximum FCN so far (86348.3) to force MIGRAD to back out of this region. Error log follows.
Parameter values: cHDD=3.22694 cHl3=-7.04051 cHq3=0.54016
RooFit::Detail::RooNormalizedPdf::wrap_pdf_over_wrap_pdf_Int[pTV][ numerator=wrap_pdf denominator=wrap_pdf_Int[pTV] ]
getLogVal() top-level p.d.f evaluates to NaN @ numerator=wrap_pdf=-2.21831, denominator=wrap_pdf_Int[pTV]=89722.5
RooAbsMinimizerFcn: Minimized function has error status.
Returning maximum FCN so far (86348.3) to force MIGRAD to back out of this region. Error log follows.
Parameter values: cHDD=3.44258 cHl3=4.96668 cHq3=0.0273884
RooFit::Detail::RooNormalizedPdf::wrap_pdf_over_wrap_pdf_Int[pTV][ numerator=wrap_pdf denominator=wrap_pdf_Int[pTV] ]
p.d.f value is less than zero, trying to recover @ numerator=wrap_pdf=0.403042, denominator=wrap_pdf_Int[pTV]=242.998
p.d.f value is less than zero, trying to recover @ numerator=wrap_pdf=0.403042, denominator=wrap_pdf_Int[pTV]=242.998
p.d.f value is less than zero, trying to recover @ numerator=wrap_pdf=0.403042, denominator=wrap_pdf_Int[pTV]=242.998
p.d.f value is less than zero, trying to recover @ numerator=wrap_pdf=0.403042, denominator=wrap_pdf_Int[pTV]=242.998
p.d.f value is less than zero, trying to recover @ numerator=wrap_pdf=0.403042, denominator=wrap_pdf_Int[pTV]=242.998
p.d.f value is less than zero, trying to recover @ numerator=wrap_pdf=0.403042, denominator=wrap_pdf_Int[pTV]=242.998
p.d.f value is less than zero, trying to recover @ numerator=wrap_pdf=0.403042, denominator=wrap_pdf_Int[pTV]=242.998
p.d.f value is less than zero, trying to recover @ numerator=wrap_pdf=0.403042, denominator=wrap_pdf_Int[pTV]=242.998
p.d.f value is less than zero, trying to recover @ numerator=wrap_pdf=0.403042, denominator=wrap_pdf_Int[pTV]=242.998
p.d.f value is less than zero, trying to recover @ numerator=wrap_pdf=0.403042, denominator=wrap_pdf_Int[pTV]=242.998
p.d.f value is less than zero, trying to recover @ numerator=wrap_pdf=0.403042, denominator=wrap_pdf_Int[pTV]=242.998
p.d.f value is less than zero, trying to recover @ numerator=wrap_pdf=0.403042, denominator=wrap_pdf_Int[pTV]=242.998
... (remaining 14 messages suppressed)
RooAbsMinimizerFcn: Minimized function has error status.
Returning maximum FCN so far (86348.3) to force MIGRAD to back out of this region. Error log follows.
Parameter values: cHDD=-3.35967 cHl3=-9.58704 cHq3=-6.27461
RooFit::Detail::RooNormalizedPdf::wrap_pdf_over_wrap_pdf_Int[pTV][ numerator=wrap_pdf denominator=wrap_pdf_Int[pTV] ]
p.d.f value is less than zero, trying to recover @ numerator=wrap_pdf=486.968, denominator=wrap_pdf_Int[pTV]=829530
p.d.f value is less than zero, trying to recover @ numerator=wrap_pdf=486.968, denominator=wrap_pdf_Int[pTV]=829530
getLogVal() top-level p.d.f evaluates to NaN @ numerator=wrap_pdf=486.968, denominator=wrap_pdf_Int[pTV]=829530
getLogVal() top-level p.d.f evaluates to NaN @ numerator=wrap_pdf=486.968, denominator=wrap_pdf_Int[pTV]=829530
RooAbsMinimizerFcn: Minimized function has error status.
Returning maximum FCN so far (86348.3) to force MIGRAD to back out of this region. Error log follows.
Parameter values: cHDD=0.154263 cHl3=2.95902 cHq3=-2.78828
RooFit::Detail::RooNormalizedPdf::wrap_pdf_over_wrap_pdf_Int[pTV][ numerator=wrap_pdf denominator=wrap_pdf_Int[pTV] ]
getLogVal() top-level p.d.f evaluates to NaN @ numerator=wrap_pdf=59.1285, denominator=wrap_pdf_Int[pTV]=200921
RooAbsMinimizerFcn: Minimized function has error status.
Returning maximum FCN so far (86348.3) to force MIGRAD to back out of this region. Error log follows.
Parameter values: cHDD=0.338546 cHl3=0.879879 cHq3=-1.35856
RooFit::Detail::RooNormalizedPdf::wrap_pdf_over_wrap_pdf_Int[pTV][ numerator=wrap_pdf denominator=wrap_pdf_Int[pTV] ]
p.d.f value is less than zero, trying to recover @ numerator=wrap_pdf=14.3535, denominator=wrap_pdf_Int[pTV]=34082.1
p.d.f value is less than zero, trying to recover @ numerator=wrap_pdf=14.3535, denominator=wrap_pdf_Int[pTV]=34082.1
getLogVal() top-level p.d.f evaluates to NaN @ numerator=wrap_pdf=14.3535, denominator=wrap_pdf_Int[pTV]=34082.1
getLogVal() top-level p.d.f evaluates to NaN @ numerator=wrap_pdf=14.3535, denominator=wrap_pdf_Int[pTV]=34082.1
RooAbsMinimizerFcn: Minimized function has error status.
Returning maximum FCN so far (86348.3) to force MIGRAD to back out of this region. Error log follows.
Parameter values: cHDD=0.372361 cHl3=0.491134 cHq3=-0.886807
RooFit::Detail::RooNormalizedPdf::wrap_pdf_over_wrap_pdf_Int[pTV][ numerator=wrap_pdf denominator=wrap_pdf_Int[pTV] ]
getLogVal() top-level p.d.f evaluates to NaN @ numerator=wrap_pdf=5.8312, denominator=wrap_pdf_Int[pTV]=12183.6
[#1] INFO:Fitting -- RooAbsPdf::fitTo(wrap_pdf) Calculating sum-of-weights-squared correction matrix for covariance matrix
[#1] INFO:InputArguments -- RooAbsData::plotOn(pseudo_dh) INFO: dataset has non-integer weights, auto-selecting SumW2 errors instead of Poisson errors
[#1] INFO:Plotting -- RooPlot::updateFitRangeNorm: New event count of 7389.24 will supersede previous event count of 9313.81 for normalization of PDF projections