[#0] WARNING:InputArguments -- The parameter 'sigG' with range [-10, 10] of the RooGaussian 'g' exceeds the safe range of (0, inf). Advise to limit its range.
[#1] INFO:Minimization -- p.d.f. provides expected number of events, including extended term in likelihood.
[#1] INFO:Fitting -- RooAbsPdf::fitTo(model0_over_model0_Int[x]) fixing normalization set for coefficient determination to observables in data
[#1] INFO:Fitting -- using generic CPU library compiled with no vectorizations
[#1] INFO:Fitting -- Creation of NLL object took 829.093 μs
[#1] INFO:Fitting -- RooAddition::defaultErrorLevel(nll_model0_over_model0_Int[x]_sumData) Summation contains a RooNLLVar, using its error level
[#1] INFO:Minimization -- RooAbsMinimizerFcn::setOptimizeConst: activating const optimization
[#1] INFO:Minimization -- [fitFCN] No discrete parameters, performing continuous minimization only
Minuit2Minimizer: Minimize with max-calls 1000 convergence for edm < 1 strategy 1
Minuit2Minimizer : Valid minimum - status = 0
FVAL = -2388.31039421315518
Edm = 3.25299757922590944e-06
Nfcn = 60
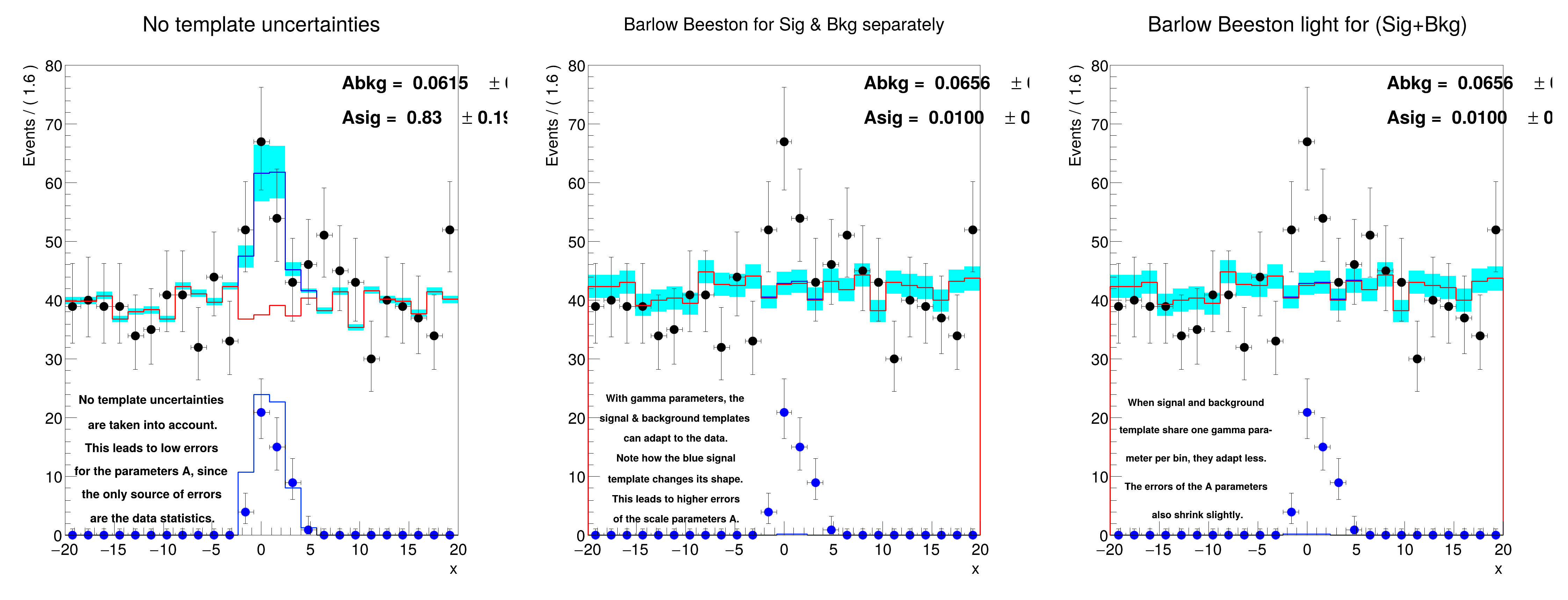
Abkg = 0.0614547 +/- 0.00211669 (limited)
Asig = 0.833778 +/- 0.1898 (limited)
[#1] INFO:Minimization -- RooAbsMinimizerFcn::setOptimizeConst: deactivating const optimization
[#1] INFO:Minimization -- p.d.f. provides expected number of events, including extended term in likelihood.
[#1] INFO:Minimization -- Including the following constraint terms in minimization: (hc_bkg,hc_sig)
[#1] INFO:Minimization -- The global observables are not defined , normalize constraints with respect to the parameters (Abkg,Asig,p_ph_bkg_gamma_bin_0,p_ph_bkg_gamma_bin_1,p_ph_bkg_gamma_bin_10,p_ph_bkg_gamma_bin_11,p_ph_bkg_gamma_bin_12,p_ph_bkg_gamma_bin_13,p_ph_bkg_gamma_bin_14,p_ph_bkg_gamma_bin_15,p_ph_bkg_gamma_bin_16,p_ph_bkg_gamma_bin_17,p_ph_bkg_gamma_bin_18,p_ph_bkg_gamma_bin_19,p_ph_bkg_gamma_bin_2,p_ph_bkg_gamma_bin_20,p_ph_bkg_gamma_bin_21,p_ph_bkg_gamma_bin_22,p_ph_bkg_gamma_bin_23,p_ph_bkg_gamma_bin_24,p_ph_bkg_gamma_bin_3,p_ph_bkg_gamma_bin_4,p_ph_bkg_gamma_bin_5,p_ph_bkg_gamma_bin_6,p_ph_bkg_gamma_bin_7,p_ph_bkg_gamma_bin_8,p_ph_bkg_gamma_bin_9,p_ph_sig_gamma_bin_0,p_ph_sig_gamma_bin_1,p_ph_sig_gamma_bin_10,p_ph_sig_gamma_bin_11,p_ph_sig_gamma_bin_12,p_ph_sig_gamma_bin_13,p_ph_sig_gamma_bin_14,p_ph_sig_gamma_bin_15,p_ph_sig_gamma_bin_16,p_ph_sig_gamma_bin_17,p_ph_sig_gamma_bin_18,p_ph_sig_gamma_bin_19,p_ph_sig_gamma_bin_2,p_ph_sig_gamma_bin_20,p_ph_sig_gamma_bin_21,p_ph_sig_gamma_bin_22,p_ph_sig_gamma_bin_23,p_ph_sig_gamma_bin_24,p_ph_sig_gamma_bin_3,p_ph_sig_gamma_bin_4,p_ph_sig_gamma_bin_5,p_ph_sig_gamma_bin_6,p_ph_sig_gamma_bin_7,p_ph_sig_gamma_bin_8,p_ph_sig_gamma_bin_9)
[#1] INFO:Fitting -- RooAbsPdf::fitTo(model1) fixing normalization set for coefficient determination to observables in data
[#1] INFO:Fitting -- Creation of NLL object took 1.63401 ms
[#1] INFO:Fitting -- RooAddition::defaultErrorLevel(nll_model1_sumData) Summation contains a RooNLLVar, using its error level
[#1] INFO:Minimization -- RooAbsMinimizerFcn::setOptimizeConst: activating const optimization
[#1] INFO:Minimization -- [fitFCN] No discrete parameters, performing continuous minimization only
Minuit2Minimizer: Minimize with max-calls 16000 convergence for edm < 1 strategy 1
RooAbsMinimizerFcn: Minimized function has error status.
Returning maximum FCN so far (10425.6) to force MIGRAD to back out of this region. Error log follows.
Parameter values: Abkg=0.024352 Asig=0.0100451 p_ph_bkg_gamma_bin_0=0.351482 p_ph_bkg_gamma_bin_1=0.352595 p_ph_bkg_gamma_bin_10=0.342683 p_ph_bkg_gamma_bin_11=0.368891 p_ph_bkg_gamma_bin_12=0.38197 p_ph_bkg_gamma_bin_13=0.365529 p_ph_bkg_gamma_bin_14=0.358098 p_ph_bkg_gamma_bin_15=0.358584 p_ph_bkg_gamma_bin_16=0.367344 p_ph_bkg_gamma_bin_17=0.356263 p_ph_bkg_gamma_bin_18=0.362211 p_ph_bkg_gamma_bin_19=0.340063 p_ph_bkg_gamma_bin_2=0.35054 p_ph_bkg_gamma_bin_20=0.352376 p_ph_bkg_gamma_bin_21=0.351589 p_ph_bkg_gamma_bin_22=0.351615 p_ph_bkg_gamma_bin_23=0.344488 p_ph_bkg_gamma_bin_24=0.365687 p_ph_bkg_gamma_bin_3=0.355197 p_ph_bkg_gamma_bin_4=0.347776 p_ph_bkg_gamma_bin_5=0.348629 p_ph_bkg_gamma_bin_6=0.357616 p_ph_bkg_gamma_bin_7=0.351061 p_ph_bkg_gamma_bin_8=0.342678 p_ph_bkg_gamma_bin_9=0.357292 p_ph_sig_gamma_bin_11=0.3339 p_ph_sig_gamma_bin_12=0.341347 p_ph_sig_gamma_bin_13=0.333434 p_ph_sig_gamma_bin_14=0.328932 p_ph_sig_gamma_bin_15=0.328666
RooAbsMinimizerFcn: Minimized function has error status.
Returning maximum FCN so far (10425.6) to force MIGRAD to back out of this region. Error log follows.
Parameter values: Abkg=0.327409 Asig=0.26768 p_ph_bkg_gamma_bin_0=0.63877 p_ph_bkg_gamma_bin_1=0.639509 p_ph_bkg_gamma_bin_10=0.632897 p_ph_bkg_gamma_bin_11=0.650254 p_ph_bkg_gamma_bin_12=0.658769 p_ph_bkg_gamma_bin_13=0.648049 p_ph_bkg_gamma_bin_14=0.643155 p_ph_bkg_gamma_bin_15=0.643476 p_ph_bkg_gamma_bin_16=0.64924 p_ph_bkg_gamma_bin_17=0.641941 p_ph_bkg_gamma_bin_18=0.645868 p_ph_bkg_gamma_bin_19=0.63114 p_ph_bkg_gamma_bin_2=0.638144 p_ph_bkg_gamma_bin_20=0.639364 p_ph_bkg_gamma_bin_21=0.638841 p_ph_bkg_gamma_bin_22=0.638858 p_ph_bkg_gamma_bin_23=0.634106 p_ph_bkg_gamma_bin_24=0.648153 p_ph_bkg_gamma_bin_3=0.641235 p_ph_bkg_gamma_bin_4=0.636302 p_ph_bkg_gamma_bin_5=0.636871 p_ph_bkg_gamma_bin_6=0.642836 p_ph_bkg_gamma_bin_7=0.63849 p_ph_bkg_gamma_bin_8=0.632894 p_ph_bkg_gamma_bin_9=0.642622 p_ph_sig_gamma_bin_11=0.626988 p_ph_sig_gamma_bin_12=0.632002 p_ph_sig_gamma_bin_13=0.626672 p_ph_sig_gamma_bin_14=0.623623 p_ph_sig_gamma_bin_15=0.623442
Minuit2Minimizer : Valid minimum - status = 0
FVAL = -2282.22793579973586
Edm = 6.50103684684927942e-05
Nfcn = 730
Abkg = 0.0613644 +/- 0.00219573 (limited)
Asig = 0.850329 +/- 0.233134 (limited)
p_ph_bkg_gamma_bin_0 = 0.998158 +/- 0.0370021 (limited)
p_ph_bkg_gamma_bin_1 = 1.0004 +/- 0.0370335 (limited)
p_ph_bkg_gamma_bin_10 = 0.980402 +/- 0.0356998 (limited)
p_ph_bkg_gamma_bin_11 = 1.00613 +/- 0.0401139 (limited)
p_ph_bkg_gamma_bin_12 = 1.00446 +/- 0.0453421 (limited)
p_ph_bkg_gamma_bin_13 = 0.991203 +/- 0.0395193 (limited)
p_ph_bkg_gamma_bin_14 = 0.996054 +/- 0.0385246 (limited)
p_ph_bkg_gamma_bin_15 = 1.00908 +/- 0.0369959 (limited)
p_ph_bkg_gamma_bin_16 = 1.03008 +/- 0.0382325 (limited)
p_ph_bkg_gamma_bin_17 = 1.00779 +/- 0.0364465 (limited)
p_ph_bkg_gamma_bin_18 = 1.01976 +/- 0.0396035 (limited)
p_ph_bkg_gamma_bin_19 = 0.975108 +/- 0.0359113 (limited)
p_ph_bkg_gamma_bin_2 = 0.996261 +/- 0.0365836 (limited)
p_ph_bkg_gamma_bin_20 = 0.999962 +/- 0.0369391 (limited)
p_ph_bkg_gamma_bin_21 = 0.998374 +/- 0.0370495 (limited)
p_ph_bkg_gamma_bin_22 = 0.998427 +/- 0.0380223 (limited)
p_ph_bkg_gamma_bin_23 = 0.984048 +/- 0.0361184 (limited)
p_ph_bkg_gamma_bin_24 = 1.02675 +/- 0.0373117 (limited)
p_ph_bkg_gamma_bin_3 = 1.00565 +/- 0.0386186 (limited)
p_ph_bkg_gamma_bin_4 = 0.990685 +/- 0.0377676 (limited)
p_ph_bkg_gamma_bin_5 = 0.992405 +/- 0.0376505 (limited)
p_ph_bkg_gamma_bin_6 = 1.01052 +/- 0.038689 (limited)
p_ph_bkg_gamma_bin_7 = 0.99731 +/- 0.0359322 (limited)
p_ph_bkg_gamma_bin_8 = 0.980392 +/- 0.0362349 (limited)
p_ph_bkg_gamma_bin_9 = 1.00987 +/- 0.0372551 (limited)
p_ph_sig_gamma_bin_11 = 1.09457 +/- 0.29733 (limited)
p_ph_sig_gamma_bin_12 = 1.07037 +/- 0.213545 (limited)
p_ph_sig_gamma_bin_13 = 0.890724 +/- 0.195929 (limited)
p_ph_sig_gamma_bin_14 = 0.947374 +/- 0.309135 (limited)
p_ph_sig_gamma_bin_15 = 1.14132 +/- 0.81204 (limited)
[#1] INFO:Minimization -- RooAbsMinimizerFcn::setOptimizeConst: deactivating const optimization
[#1] INFO:Minimization -- p.d.f. provides expected number of events, including extended term in likelihood.
[#1] INFO:Minimization -- Including the following constraint terms in minimization: (hc_sigbkg)
[#1] INFO:Minimization -- The global observables are not defined , normalize constraints with respect to the parameters (Abkg,Asig,p_ph_sig2_gamma_bin_0,p_ph_sig2_gamma_bin_1,p_ph_sig2_gamma_bin_10,p_ph_sig2_gamma_bin_11,p_ph_sig2_gamma_bin_12,p_ph_sig2_gamma_bin_13,p_ph_sig2_gamma_bin_14,p_ph_sig2_gamma_bin_15,p_ph_sig2_gamma_bin_16,p_ph_sig2_gamma_bin_17,p_ph_sig2_gamma_bin_18,p_ph_sig2_gamma_bin_19,p_ph_sig2_gamma_bin_2,p_ph_sig2_gamma_bin_20,p_ph_sig2_gamma_bin_21,p_ph_sig2_gamma_bin_22,p_ph_sig2_gamma_bin_23,p_ph_sig2_gamma_bin_24,p_ph_sig2_gamma_bin_3,p_ph_sig2_gamma_bin_4,p_ph_sig2_gamma_bin_5,p_ph_sig2_gamma_bin_6,p_ph_sig2_gamma_bin_7,p_ph_sig2_gamma_bin_8,p_ph_sig2_gamma_bin_9)
[#1] INFO:Fitting -- RooAbsPdf::fitTo(model2) fixing normalization set for coefficient determination to observables in data
[#1] INFO:Fitting -- Creation of NLL object took 800.243 μs
[#1] INFO:Fitting -- RooAddition::defaultErrorLevel(nll_model2_sumData) Summation contains a RooNLLVar, using its error level
[#1] INFO:Minimization -- RooAbsMinimizerFcn::setOptimizeConst: activating const optimization
[#1] INFO:Minimization -- [fitFCN] No discrete parameters, performing continuous minimization only
Minuit2Minimizer: Minimize with max-calls 13500 convergence for edm < 1 strategy 1
RooAbsMinimizerFcn: Minimized function has error status.
Returning maximum FCN so far (10415.9) to force MIGRAD to back out of this region. Error log follows.
Parameter values: Abkg=1 Asig=1 p_ph_sig2_gamma_bin_0=7.28945 p_ph_sig2_gamma_bin_1=1 p_ph_sig2_gamma_bin_10=1 p_ph_sig2_gamma_bin_11=1 p_ph_sig2_gamma_bin_12=1 p_ph_sig2_gamma_bin_13=1 p_ph_sig2_gamma_bin_14=1 p_ph_sig2_gamma_bin_15=1 p_ph_sig2_gamma_bin_16=1 p_ph_sig2_gamma_bin_17=1 p_ph_sig2_gamma_bin_18=1 p_ph_sig2_gamma_bin_19=1 p_ph_sig2_gamma_bin_2=1 p_ph_sig2_gamma_bin_20=1 p_ph_sig2_gamma_bin_21=1 p_ph_sig2_gamma_bin_22=1 p_ph_sig2_gamma_bin_23=1 p_ph_sig2_gamma_bin_24=1 p_ph_sig2_gamma_bin_3=1 p_ph_sig2_gamma_bin_4=1 p_ph_sig2_gamma_bin_5=1 p_ph_sig2_gamma_bin_6=1 p_ph_sig2_gamma_bin_7=1 p_ph_sig2_gamma_bin_8=1 p_ph_sig2_gamma_bin_9=1
RooAbsMinimizerFcn: Minimized function has error status.
Returning maximum FCN so far (10415.9) to force MIGRAD to back out of this region. Error log follows.
Parameter values: Abkg=1 Asig=1 p_ph_sig2_gamma_bin_0=1 p_ph_sig2_gamma_bin_1=7.28945 p_ph_sig2_gamma_bin_10=1 p_ph_sig2_gamma_bin_11=1 p_ph_sig2_gamma_bin_12=1 p_ph_sig2_gamma_bin_13=1 p_ph_sig2_gamma_bin_14=1 p_ph_sig2_gamma_bin_15=1 p_ph_sig2_gamma_bin_16=1 p_ph_sig2_gamma_bin_17=1 p_ph_sig2_gamma_bin_18=1 p_ph_sig2_gamma_bin_19=1 p_ph_sig2_gamma_bin_2=1 p_ph_sig2_gamma_bin_20=1 p_ph_sig2_gamma_bin_21=1 p_ph_sig2_gamma_bin_22=1 p_ph_sig2_gamma_bin_23=1 p_ph_sig2_gamma_bin_24=1 p_ph_sig2_gamma_bin_3=1 p_ph_sig2_gamma_bin_4=1 p_ph_sig2_gamma_bin_5=1 p_ph_sig2_gamma_bin_6=1 p_ph_sig2_gamma_bin_7=1 p_ph_sig2_gamma_bin_8=1 p_ph_sig2_gamma_bin_9=1
RooAbsMinimizerFcn: Minimized function has error status.
Returning maximum FCN so far (10415.9) to force MIGRAD to back out of this region. Error log follows.
Parameter values: Abkg=1 Asig=1 p_ph_sig2_gamma_bin_0=1 p_ph_sig2_gamma_bin_1=1 p_ph_sig2_gamma_bin_10=7.28945 p_ph_sig2_gamma_bin_11=1 p_ph_sig2_gamma_bin_12=1 p_ph_sig2_gamma_bin_13=1 p_ph_sig2_gamma_bin_14=1 p_ph_sig2_gamma_bin_15=1 p_ph_sig2_gamma_bin_16=1 p_ph_sig2_gamma_bin_17=1 p_ph_sig2_gamma_bin_18=1 p_ph_sig2_gamma_bin_19=1 p_ph_sig2_gamma_bin_2=1 p_ph_sig2_gamma_bin_20=1 p_ph_sig2_gamma_bin_21=1 p_ph_sig2_gamma_bin_22=1 p_ph_sig2_gamma_bin_23=1 p_ph_sig2_gamma_bin_24=1 p_ph_sig2_gamma_bin_3=1 p_ph_sig2_gamma_bin_4=1 p_ph_sig2_gamma_bin_5=1 p_ph_sig2_gamma_bin_6=1 p_ph_sig2_gamma_bin_7=1 p_ph_sig2_gamma_bin_8=1 p_ph_sig2_gamma_bin_9=1
RooAbsMinimizerFcn: Minimized function has error status.
Returning maximum FCN so far (10415.9) to force MIGRAD to back out of this region. Error log follows.
Parameter values: Abkg=1 Asig=1 p_ph_sig2_gamma_bin_0=1 p_ph_sig2_gamma_bin_1=1 p_ph_sig2_gamma_bin_10=1 p_ph_sig2_gamma_bin_11=1 p_ph_sig2_gamma_bin_12=1 p_ph_sig2_gamma_bin_13=1 p_ph_sig2_gamma_bin_14=1 p_ph_sig2_gamma_bin_15=1 p_ph_sig2_gamma_bin_16=7.28945 p_ph_sig2_gamma_bin_17=1 p_ph_sig2_gamma_bin_18=1 p_ph_sig2_gamma_bin_19=1 p_ph_sig2_gamma_bin_2=1 p_ph_sig2_gamma_bin_20=1 p_ph_sig2_gamma_bin_21=1 p_ph_sig2_gamma_bin_22=1 p_ph_sig2_gamma_bin_23=1 p_ph_sig2_gamma_bin_24=1 p_ph_sig2_gamma_bin_3=1 p_ph_sig2_gamma_bin_4=1 p_ph_sig2_gamma_bin_5=1 p_ph_sig2_gamma_bin_6=1 p_ph_sig2_gamma_bin_7=1 p_ph_sig2_gamma_bin_8=1 p_ph_sig2_gamma_bin_9=1
RooAbsMinimizerFcn: Minimized function has error status.
Returning maximum FCN so far (10415.9) to force MIGRAD to back out of this region. Error log follows.
Parameter values: Abkg=1 Asig=1 p_ph_sig2_gamma_bin_0=1 p_ph_sig2_gamma_bin_1=1 p_ph_sig2_gamma_bin_10=1 p_ph_sig2_gamma_bin_11=1 p_ph_sig2_gamma_bin_12=1 p_ph_sig2_gamma_bin_13=1 p_ph_sig2_gamma_bin_14=1 p_ph_sig2_gamma_bin_15=1 p_ph_sig2_gamma_bin_16=1 p_ph_sig2_gamma_bin_17=7.28945 p_ph_sig2_gamma_bin_18=1 p_ph_sig2_gamma_bin_19=1 p_ph_sig2_gamma_bin_2=1 p_ph_sig2_gamma_bin_20=1 p_ph_sig2_gamma_bin_21=1 p_ph_sig2_gamma_bin_22=1 p_ph_sig2_gamma_bin_23=1 p_ph_sig2_gamma_bin_24=1 p_ph_sig2_gamma_bin_3=1 p_ph_sig2_gamma_bin_4=1 p_ph_sig2_gamma_bin_5=1 p_ph_sig2_gamma_bin_6=1 p_ph_sig2_gamma_bin_7=1 p_ph_sig2_gamma_bin_8=1 p_ph_sig2_gamma_bin_9=1
RooAbsMinimizerFcn: Minimized function has error status.
Returning maximum FCN so far (10415.9) to force MIGRAD to back out of this region. Error log follows.
Parameter values: Abkg=1 Asig=1 p_ph_sig2_gamma_bin_0=1 p_ph_sig2_gamma_bin_1=1 p_ph_sig2_gamma_bin_10=1 p_ph_sig2_gamma_bin_11=1 p_ph_sig2_gamma_bin_12=1 p_ph_sig2_gamma_bin_13=1 p_ph_sig2_gamma_bin_14=1 p_ph_sig2_gamma_bin_15=1 p_ph_sig2_gamma_bin_16=1 p_ph_sig2_gamma_bin_17=1 p_ph_sig2_gamma_bin_18=7.28945 p_ph_sig2_gamma_bin_19=1 p_ph_sig2_gamma_bin_2=1 p_ph_sig2_gamma_bin_20=1 p_ph_sig2_gamma_bin_21=1 p_ph_sig2_gamma_bin_22=1 p_ph_sig2_gamma_bin_23=1 p_ph_sig2_gamma_bin_24=1 p_ph_sig2_gamma_bin_3=1 p_ph_sig2_gamma_bin_4=1 p_ph_sig2_gamma_bin_5=1 p_ph_sig2_gamma_bin_6=1 p_ph_sig2_gamma_bin_7=1 p_ph_sig2_gamma_bin_8=1 p_ph_sig2_gamma_bin_9=1
RooAbsMinimizerFcn: Minimized function has error status.
Returning maximum FCN so far (10415.9) to force MIGRAD to back out of this region. Error log follows.
Parameter values: Abkg=1 Asig=1 p_ph_sig2_gamma_bin_0=1 p_ph_sig2_gamma_bin_1=1 p_ph_sig2_gamma_bin_10=1 p_ph_sig2_gamma_bin_11=1 p_ph_sig2_gamma_bin_12=1 p_ph_sig2_gamma_bin_13=1 p_ph_sig2_gamma_bin_14=1 p_ph_sig2_gamma_bin_15=1 p_ph_sig2_gamma_bin_16=1 p_ph_sig2_gamma_bin_17=1 p_ph_sig2_gamma_bin_18=1 p_ph_sig2_gamma_bin_19=7.28945 p_ph_sig2_gamma_bin_2=1 p_ph_sig2_gamma_bin_20=1 p_ph_sig2_gamma_bin_21=1 p_ph_sig2_gamma_bin_22=1 p_ph_sig2_gamma_bin_23=1 p_ph_sig2_gamma_bin_24=1 p_ph_sig2_gamma_bin_3=1 p_ph_sig2_gamma_bin_4=1 p_ph_sig2_gamma_bin_5=1 p_ph_sig2_gamma_bin_6=1 p_ph_sig2_gamma_bin_7=1 p_ph_sig2_gamma_bin_8=1 p_ph_sig2_gamma_bin_9=1
RooAbsMinimizerFcn: Minimized function has error status.
Returning maximum FCN so far (10415.9) to force MIGRAD to back out of this region. Error log follows.
Parameter values: Abkg=1 Asig=1 p_ph_sig2_gamma_bin_0=1 p_ph_sig2_gamma_bin_1=1 p_ph_sig2_gamma_bin_10=1 p_ph_sig2_gamma_bin_11=1 p_ph_sig2_gamma_bin_12=1 p_ph_sig2_gamma_bin_13=1 p_ph_sig2_gamma_bin_14=1 p_ph_sig2_gamma_bin_15=1 p_ph_sig2_gamma_bin_16=1 p_ph_sig2_gamma_bin_17=1 p_ph_sig2_gamma_bin_18=1 p_ph_sig2_gamma_bin_19=1 p_ph_sig2_gamma_bin_2=7.28945 p_ph_sig2_gamma_bin_20=1 p_ph_sig2_gamma_bin_21=1 p_ph_sig2_gamma_bin_22=1 p_ph_sig2_gamma_bin_23=1 p_ph_sig2_gamma_bin_24=1 p_ph_sig2_gamma_bin_3=1 p_ph_sig2_gamma_bin_4=1 p_ph_sig2_gamma_bin_5=1 p_ph_sig2_gamma_bin_6=1 p_ph_sig2_gamma_bin_7=1 p_ph_sig2_gamma_bin_8=1 p_ph_sig2_gamma_bin_9=1
RooAbsMinimizerFcn: Minimized function has error status.
Returning maximum FCN so far (10415.9) to force MIGRAD to back out of this region. Error log follows.
Parameter values: Abkg=1 Asig=1 p_ph_sig2_gamma_bin_0=1 p_ph_sig2_gamma_bin_1=1 p_ph_sig2_gamma_bin_10=1 p_ph_sig2_gamma_bin_11=1 p_ph_sig2_gamma_bin_12=1 p_ph_sig2_gamma_bin_13=1 p_ph_sig2_gamma_bin_14=1 p_ph_sig2_gamma_bin_15=1 p_ph_sig2_gamma_bin_16=1 p_ph_sig2_gamma_bin_17=1 p_ph_sig2_gamma_bin_18=1 p_ph_sig2_gamma_bin_19=1 p_ph_sig2_gamma_bin_2=1 p_ph_sig2_gamma_bin_20=7.28945 p_ph_sig2_gamma_bin_21=1 p_ph_sig2_gamma_bin_22=1 p_ph_sig2_gamma_bin_23=1 p_ph_sig2_gamma_bin_24=1 p_ph_sig2_gamma_bin_3=1 p_ph_sig2_gamma_bin_4=1 p_ph_sig2_gamma_bin_5=1 p_ph_sig2_gamma_bin_6=1 p_ph_sig2_gamma_bin_7=1 p_ph_sig2_gamma_bin_8=1 p_ph_sig2_gamma_bin_9=1
RooAbsMinimizerFcn: Minimized function has error status.
Returning maximum FCN so far (10415.9) to force MIGRAD to back out of this region. Error log follows.
Parameter values: Abkg=1 Asig=1 p_ph_sig2_gamma_bin_0=1 p_ph_sig2_gamma_bin_1=1 p_ph_sig2_gamma_bin_10=1 p_ph_sig2_gamma_bin_11=1 p_ph_sig2_gamma_bin_12=1 p_ph_sig2_gamma_bin_13=1 p_ph_sig2_gamma_bin_14=1 p_ph_sig2_gamma_bin_15=1 p_ph_sig2_gamma_bin_16=1 p_ph_sig2_gamma_bin_17=1 p_ph_sig2_gamma_bin_18=1 p_ph_sig2_gamma_bin_19=1 p_ph_sig2_gamma_bin_2=1 p_ph_sig2_gamma_bin_20=1 p_ph_sig2_gamma_bin_21=7.28945 p_ph_sig2_gamma_bin_22=1 p_ph_sig2_gamma_bin_23=1 p_ph_sig2_gamma_bin_24=1 p_ph_sig2_gamma_bin_3=1 p_ph_sig2_gamma_bin_4=1 p_ph_sig2_gamma_bin_5=1 p_ph_sig2_gamma_bin_6=1 p_ph_sig2_gamma_bin_7=1 p_ph_sig2_gamma_bin_8=1 p_ph_sig2_gamma_bin_9=1
RooAbsMinimizerFcn: Minimized function has error status.
Returning maximum FCN so far (10415.9) to force MIGRAD to back out of this region. Error log follows.
Parameter values: Abkg=1 Asig=1 p_ph_sig2_gamma_bin_0=1 p_ph_sig2_gamma_bin_1=1 p_ph_sig2_gamma_bin_10=1 p_ph_sig2_gamma_bin_11=1 p_ph_sig2_gamma_bin_12=1 p_ph_sig2_gamma_bin_13=1 p_ph_sig2_gamma_bin_14=1 p_ph_sig2_gamma_bin_15=1 p_ph_sig2_gamma_bin_16=1 p_ph_sig2_gamma_bin_17=1 p_ph_sig2_gamma_bin_18=1 p_ph_sig2_gamma_bin_19=1 p_ph_sig2_gamma_bin_2=1 p_ph_sig2_gamma_bin_20=1 p_ph_sig2_gamma_bin_21=1 p_ph_sig2_gamma_bin_22=7.28945 p_ph_sig2_gamma_bin_23=1 p_ph_sig2_gamma_bin_24=1 p_ph_sig2_gamma_bin_3=1 p_ph_sig2_gamma_bin_4=1 p_ph_sig2_gamma_bin_5=1 p_ph_sig2_gamma_bin_6=1 p_ph_sig2_gamma_bin_7=1 p_ph_sig2_gamma_bin_8=1 p_ph_sig2_gamma_bin_9=1
RooAbsMinimizerFcn: Minimized function has error status.
Returning maximum FCN so far (10415.9) to force MIGRAD to back out of this region. Error log follows.
Parameter values: Abkg=1 Asig=1 p_ph_sig2_gamma_bin_0=1 p_ph_sig2_gamma_bin_1=1 p_ph_sig2_gamma_bin_10=1 p_ph_sig2_gamma_bin_11=1 p_ph_sig2_gamma_bin_12=1 p_ph_sig2_gamma_bin_13=1 p_ph_sig2_gamma_bin_14=1 p_ph_sig2_gamma_bin_15=1 p_ph_sig2_gamma_bin_16=1 p_ph_sig2_gamma_bin_17=1 p_ph_sig2_gamma_bin_18=1 p_ph_sig2_gamma_bin_19=1 p_ph_sig2_gamma_bin_2=1 p_ph_sig2_gamma_bin_20=1 p_ph_sig2_gamma_bin_21=1 p_ph_sig2_gamma_bin_22=1 p_ph_sig2_gamma_bin_23=7.28945 p_ph_sig2_gamma_bin_24=1 p_ph_sig2_gamma_bin_3=1 p_ph_sig2_gamma_bin_4=1 p_ph_sig2_gamma_bin_5=1 p_ph_sig2_gamma_bin_6=1 p_ph_sig2_gamma_bin_7=1 p_ph_sig2_gamma_bin_8=1 p_ph_sig2_gamma_bin_9=1
RooAbsMinimizerFcn: Minimized function has error status.
Returning maximum FCN so far (10415.9) to force MIGRAD to back out of this region. Error log follows.
Parameter values: Abkg=1 Asig=1 p_ph_sig2_gamma_bin_0=1 p_ph_sig2_gamma_bin_1=1 p_ph_sig2_gamma_bin_10=1 p_ph_sig2_gamma_bin_11=1 p_ph_sig2_gamma_bin_12=1 p_ph_sig2_gamma_bin_13=1 p_ph_sig2_gamma_bin_14=1 p_ph_sig2_gamma_bin_15=1 p_ph_sig2_gamma_bin_16=1 p_ph_sig2_gamma_bin_17=1 p_ph_sig2_gamma_bin_18=1 p_ph_sig2_gamma_bin_19=1 p_ph_sig2_gamma_bin_2=1 p_ph_sig2_gamma_bin_20=1 p_ph_sig2_gamma_bin_21=1 p_ph_sig2_gamma_bin_22=1 p_ph_sig2_gamma_bin_23=1 p_ph_sig2_gamma_bin_24=7.28945 p_ph_sig2_gamma_bin_3=1 p_ph_sig2_gamma_bin_4=1 p_ph_sig2_gamma_bin_5=1 p_ph_sig2_gamma_bin_6=1 p_ph_sig2_gamma_bin_7=1 p_ph_sig2_gamma_bin_8=1 p_ph_sig2_gamma_bin_9=1
RooAbsMinimizerFcn: Minimized function has error status.
Returning maximum FCN so far (10415.9) to force MIGRAD to back out of this region. Error log follows.
Parameter values: Abkg=1 Asig=1 p_ph_sig2_gamma_bin_0=1 p_ph_sig2_gamma_bin_1=1 p_ph_sig2_gamma_bin_10=1 p_ph_sig2_gamma_bin_11=1 p_ph_sig2_gamma_bin_12=1 p_ph_sig2_gamma_bin_13=1 p_ph_sig2_gamma_bin_14=1 p_ph_sig2_gamma_bin_15=1 p_ph_sig2_gamma_bin_16=1 p_ph_sig2_gamma_bin_17=1 p_ph_sig2_gamma_bin_18=1 p_ph_sig2_gamma_bin_19=1 p_ph_sig2_gamma_bin_2=1 p_ph_sig2_gamma_bin_20=1 p_ph_sig2_gamma_bin_21=1 p_ph_sig2_gamma_bin_22=1 p_ph_sig2_gamma_bin_23=1 p_ph_sig2_gamma_bin_24=1 p_ph_sig2_gamma_bin_3=7.28945 p_ph_sig2_gamma_bin_4=1 p_ph_sig2_gamma_bin_5=1 p_ph_sig2_gamma_bin_6=1 p_ph_sig2_gamma_bin_7=1 p_ph_sig2_gamma_bin_8=1 p_ph_sig2_gamma_bin_9=1
RooAbsMinimizerFcn: Minimized function has error status.
Returning maximum FCN so far (10415.9) to force MIGRAD to back out of this region. Error log follows.
Parameter values: Abkg=1 Asig=1 p_ph_sig2_gamma_bin_0=1 p_ph_sig2_gamma_bin_1=1 p_ph_sig2_gamma_bin_10=1 p_ph_sig2_gamma_bin_11=1 p_ph_sig2_gamma_bin_12=1 p_ph_sig2_gamma_bin_13=1 p_ph_sig2_gamma_bin_14=1 p_ph_sig2_gamma_bin_15=1 p_ph_sig2_gamma_bin_16=1 p_ph_sig2_gamma_bin_17=1 p_ph_sig2_gamma_bin_18=1 p_ph_sig2_gamma_bin_19=1 p_ph_sig2_gamma_bin_2=1 p_ph_sig2_gamma_bin_20=1 p_ph_sig2_gamma_bin_21=1 p_ph_sig2_gamma_bin_22=1 p_ph_sig2_gamma_bin_23=1 p_ph_sig2_gamma_bin_24=1 p_ph_sig2_gamma_bin_3=1 p_ph_sig2_gamma_bin_4=7.28945 p_ph_sig2_gamma_bin_5=1 p_ph_sig2_gamma_bin_6=1 p_ph_sig2_gamma_bin_7=1 p_ph_sig2_gamma_bin_8=1 p_ph_sig2_gamma_bin_9=1
RooAbsMinimizerFcn: Minimized function has error status.
Returning maximum FCN so far (10415.9) to force MIGRAD to back out of this region. Error log follows.
Parameter values: Abkg=1 Asig=1 p_ph_sig2_gamma_bin_0=1 p_ph_sig2_gamma_bin_1=1 p_ph_sig2_gamma_bin_10=1 p_ph_sig2_gamma_bin_11=1 p_ph_sig2_gamma_bin_12=1 p_ph_sig2_gamma_bin_13=1 p_ph_sig2_gamma_bin_14=1 p_ph_sig2_gamma_bin_15=1 p_ph_sig2_gamma_bin_16=1 p_ph_sig2_gamma_bin_17=1 p_ph_sig2_gamma_bin_18=1 p_ph_sig2_gamma_bin_19=1 p_ph_sig2_gamma_bin_2=1 p_ph_sig2_gamma_bin_20=1 p_ph_sig2_gamma_bin_21=1 p_ph_sig2_gamma_bin_22=1 p_ph_sig2_gamma_bin_23=1 p_ph_sig2_gamma_bin_24=1 p_ph_sig2_gamma_bin_3=1 p_ph_sig2_gamma_bin_4=1 p_ph_sig2_gamma_bin_5=7.28945 p_ph_sig2_gamma_bin_6=1 p_ph_sig2_gamma_bin_7=1 p_ph_sig2_gamma_bin_8=1 p_ph_sig2_gamma_bin_9=1
RooAbsMinimizerFcn: Minimized function has error status.
Returning maximum FCN so far (10415.9) to force MIGRAD to back out of this region. Error log follows.
Parameter values: Abkg=1 Asig=1 p_ph_sig2_gamma_bin_0=1 p_ph_sig2_gamma_bin_1=1 p_ph_sig2_gamma_bin_10=1 p_ph_sig2_gamma_bin_11=1 p_ph_sig2_gamma_bin_12=1 p_ph_sig2_gamma_bin_13=1 p_ph_sig2_gamma_bin_14=1 p_ph_sig2_gamma_bin_15=1 p_ph_sig2_gamma_bin_16=1 p_ph_sig2_gamma_bin_17=1 p_ph_sig2_gamma_bin_18=1 p_ph_sig2_gamma_bin_19=1 p_ph_sig2_gamma_bin_2=1 p_ph_sig2_gamma_bin_20=1 p_ph_sig2_gamma_bin_21=1 p_ph_sig2_gamma_bin_22=1 p_ph_sig2_gamma_bin_23=1 p_ph_sig2_gamma_bin_24=1 p_ph_sig2_gamma_bin_3=1 p_ph_sig2_gamma_bin_4=1 p_ph_sig2_gamma_bin_5=1 p_ph_sig2_gamma_bin_6=7.28945 p_ph_sig2_gamma_bin_7=1 p_ph_sig2_gamma_bin_8=1 p_ph_sig2_gamma_bin_9=1
RooAbsMinimizerFcn: Minimized function has error status.
Returning maximum FCN so far (10415.9) to force MIGRAD to back out of this region. Error log follows.
Parameter values: Abkg=1 Asig=1 p_ph_sig2_gamma_bin_0=1 p_ph_sig2_gamma_bin_1=1 p_ph_sig2_gamma_bin_10=1 p_ph_sig2_gamma_bin_11=1 p_ph_sig2_gamma_bin_12=1 p_ph_sig2_gamma_bin_13=1 p_ph_sig2_gamma_bin_14=1 p_ph_sig2_gamma_bin_15=1 p_ph_sig2_gamma_bin_16=1 p_ph_sig2_gamma_bin_17=1 p_ph_sig2_gamma_bin_18=1 p_ph_sig2_gamma_bin_19=1 p_ph_sig2_gamma_bin_2=1 p_ph_sig2_gamma_bin_20=1 p_ph_sig2_gamma_bin_21=1 p_ph_sig2_gamma_bin_22=1 p_ph_sig2_gamma_bin_23=1 p_ph_sig2_gamma_bin_24=1 p_ph_sig2_gamma_bin_3=1 p_ph_sig2_gamma_bin_4=1 p_ph_sig2_gamma_bin_5=1 p_ph_sig2_gamma_bin_6=1 p_ph_sig2_gamma_bin_7=7.28945 p_ph_sig2_gamma_bin_8=1 p_ph_sig2_gamma_bin_9=1
RooAbsMinimizerFcn: Minimized function has error status.
Returning maximum FCN so far (10415.9) to force MIGRAD to back out of this region. Error log follows.
Parameter values: Abkg=1 Asig=1 p_ph_sig2_gamma_bin_0=1 p_ph_sig2_gamma_bin_1=1 p_ph_sig2_gamma_bin_10=1 p_ph_sig2_gamma_bin_11=1 p_ph_sig2_gamma_bin_12=1 p_ph_sig2_gamma_bin_13=1 p_ph_sig2_gamma_bin_14=1 p_ph_sig2_gamma_bin_15=1 p_ph_sig2_gamma_bin_16=1 p_ph_sig2_gamma_bin_17=1 p_ph_sig2_gamma_bin_18=1 p_ph_sig2_gamma_bin_19=1 p_ph_sig2_gamma_bin_2=1 p_ph_sig2_gamma_bin_20=1 p_ph_sig2_gamma_bin_21=1 p_ph_sig2_gamma_bin_22=1 p_ph_sig2_gamma_bin_23=1 p_ph_sig2_gamma_bin_24=1 p_ph_sig2_gamma_bin_3=1 p_ph_sig2_gamma_bin_4=1 p_ph_sig2_gamma_bin_5=1 p_ph_sig2_gamma_bin_6=1 p_ph_sig2_gamma_bin_7=1 p_ph_sig2_gamma_bin_8=7.28945 p_ph_sig2_gamma_bin_9=1
RooAbsMinimizerFcn: Minimized function has error status.
Returning maximum FCN so far (10415.9) to force MIGRAD to back out of this region. Error log follows.
Parameter values: Abkg=1 Asig=1 p_ph_sig2_gamma_bin_0=1 p_ph_sig2_gamma_bin_1=1 p_ph_sig2_gamma_bin_10=1 p_ph_sig2_gamma_bin_11=1 p_ph_sig2_gamma_bin_12=1 p_ph_sig2_gamma_bin_13=1 p_ph_sig2_gamma_bin_14=1 p_ph_sig2_gamma_bin_15=1 p_ph_sig2_gamma_bin_16=1 p_ph_sig2_gamma_bin_17=1 p_ph_sig2_gamma_bin_18=1 p_ph_sig2_gamma_bin_19=1 p_ph_sig2_gamma_bin_2=1 p_ph_sig2_gamma_bin_20=1 p_ph_sig2_gamma_bin_21=1 p_ph_sig2_gamma_bin_22=1 p_ph_sig2_gamma_bin_23=1 p_ph_sig2_gamma_bin_24=1 p_ph_sig2_gamma_bin_3=1 p_ph_sig2_gamma_bin_4=1 p_ph_sig2_gamma_bin_5=1 p_ph_sig2_gamma_bin_6=1 p_ph_sig2_gamma_bin_7=1 p_ph_sig2_gamma_bin_8=1 p_ph_sig2_gamma_bin_9=7.28945
RooAbsMinimizerFcn: Minimized function has error status.
Returning maximum FCN so far (10415.9) to force MIGRAD to back out of this region. Error log follows.
Parameter values: Abkg=0.024352 Asig=0.0100451 p_ph_sig2_gamma_bin_0=0.351536 p_ph_sig2_gamma_bin_1=0.352649 p_ph_sig2_gamma_bin_10=0.342736 p_ph_sig2_gamma_bin_11=0.36962 p_ph_sig2_gamma_bin_12=0.383863 p_ph_sig2_gamma_bin_13=0.366897 p_ph_sig2_gamma_bin_14=0.358549 p_ph_sig2_gamma_bin_15=0.358659 p_ph_sig2_gamma_bin_16=0.3674 p_ph_sig2_gamma_bin_17=0.356318 p_ph_sig2_gamma_bin_18=0.362267 p_ph_sig2_gamma_bin_19=0.340116 p_ph_sig2_gamma_bin_2=0.350595 p_ph_sig2_gamma_bin_20=0.352431 p_ph_sig2_gamma_bin_21=0.351643 p_ph_sig2_gamma_bin_22=0.35167 p_ph_sig2_gamma_bin_23=0.344542 p_ph_sig2_gamma_bin_24=0.365744 p_ph_sig2_gamma_bin_3=0.355252 p_ph_sig2_gamma_bin_4=0.34783 p_ph_sig2_gamma_bin_5=0.348683 p_ph_sig2_gamma_bin_6=0.357671 p_ph_sig2_gamma_bin_7=0.351115 p_ph_sig2_gamma_bin_8=0.342731 p_ph_sig2_gamma_bin_9=0.357348
RooAbsMinimizerFcn: Minimized function has error status.
Returning maximum FCN so far (10415.9) to force MIGRAD to back out of this region. Error log follows.
Parameter values: Abkg=0.327407 Asig=0.267678 p_ph_sig2_gamma_bin_0=0.638805 p_ph_sig2_gamma_bin_1=0.639545 p_ph_sig2_gamma_bin_10=0.632932 p_ph_sig2_gamma_bin_11=0.650729 p_ph_sig2_gamma_bin_12=0.659993 p_ph_sig2_gamma_bin_13=0.648946 p_ph_sig2_gamma_bin_14=0.643452 p_ph_sig2_gamma_bin_15=0.643524 p_ph_sig2_gamma_bin_16=0.649276 p_ph_sig2_gamma_bin_17=0.641977 p_ph_sig2_gamma_bin_18=0.645904 p_ph_sig2_gamma_bin_19=0.631174 p_ph_sig2_gamma_bin_2=0.638179 p_ph_sig2_gamma_bin_20=0.6394 p_ph_sig2_gamma_bin_21=0.638876 p_ph_sig2_gamma_bin_22=0.638894 p_ph_sig2_gamma_bin_23=0.634141 p_ph_sig2_gamma_bin_24=0.648189 p_ph_sig2_gamma_bin_3=0.641271 p_ph_sig2_gamma_bin_4=0.636338 p_ph_sig2_gamma_bin_5=0.636906 p_ph_sig2_gamma_bin_6=0.642872 p_ph_sig2_gamma_bin_7=0.638525 p_ph_sig2_gamma_bin_8=0.632929 p_ph_sig2_gamma_bin_9=0.642658
Minuit2Minimizer : Valid minimum - status = 0
FVAL = -2291.45108203063683
Edm = 0.000217118570047712786
Nfcn = 1108
Abkg = 0.0614871 +/- 0.00221998 (limited)
Asig = 0.834474 +/- 0.202991 (limited)
p_ph_sig2_gamma_bin_0 = 0.997805 +/- 0.0474115 (limited)
p_ph_sig2_gamma_bin_1 = 1.00004 +/- 0.0474649 (limited)
p_ph_sig2_gamma_bin_10 = 0.980078 +/- 0.0456101 (limited)
p_ph_sig2_gamma_bin_11 = 1.01069 +/- 0.0488754 (limited)
p_ph_sig2_gamma_bin_12 = 1.01199 +/- 0.0483997 (limited)
p_ph_sig2_gamma_bin_13 = 0.983813 +/- 0.0467055 (limited)
p_ph_sig2_gamma_bin_14 = 0.994769 +/- 0.0483032 (limited)
p_ph_sig2_gamma_bin_15 = 1.0095 +/- 0.0473249 (limited)
p_ph_sig2_gamma_bin_16 = 1.02967 +/- 0.049202 (limited)
p_ph_sig2_gamma_bin_17 = 1.00742 +/- 0.0467328 (limited)
p_ph_sig2_gamma_bin_18 = 1.01937 +/- 0.0509499 (limited)
p_ph_sig2_gamma_bin_19 = 0.974793 +/- 0.0458586 (limited)
p_ph_sig2_gamma_bin_2 = 0.99591 +/- 0.0468523 (limited)
p_ph_sig2_gamma_bin_20 = 0.999605 +/- 0.0473385 (limited)
p_ph_sig2_gamma_bin_21 = 0.99802 +/- 0.0474749 (limited)
p_ph_sig2_gamma_bin_22 = 0.998073 +/- 0.0487512 (limited)
p_ph_sig2_gamma_bin_23 = 0.983718 +/- 0.0461774 (limited)
p_ph_sig2_gamma_bin_24 = 1.02635 +/- 0.0479702 (limited)
p_ph_sig2_gamma_bin_3 = 1.00528 +/- 0.0495745 (limited)
p_ph_sig2_gamma_bin_4 = 0.990344 +/- 0.0483736 (limited)
p_ph_sig2_gamma_bin_5 = 0.992062 +/- 0.0482299 (limited)
p_ph_sig2_gamma_bin_6 = 1.01014 +/- 0.0496947 (limited)
p_ph_sig2_gamma_bin_7 = 0.996957 +/- 0.0460029 (limited)
p_ph_sig2_gamma_bin_8 = 0.980068 +/- 0.0463103 (limited)
p_ph_sig2_gamma_bin_9 = 1.00949 +/- 0.0478068 (limited)
[#1] INFO:Minimization -- RooAbsMinimizerFcn::setOptimizeConst: deactivating const optimization
[#0] WARNING:InputArguments -- RooAbsReal::plotOn(model0) WARNING: argument CurveNameSuffix is duplicated
[#0] WARNING:InputArguments -- RooAbsReal::plotOn(model0) WARNING: argument CurveNameSuffix is duplicated
[#0] WARNING:InputArguments -- RooAbsReal::plotOn(model0) WARNING: argument CurveNameSuffix is duplicated
[#0] WARNING:InputArguments -- RooAbsReal::plotOn(model0) WARNING: argument CurveNameSuffix is duplicated
[#0] WARNING:InputArguments -- RooAbsReal::plotOn(model0) WARNING: argument CurveNameSuffix is duplicated
[#1] INFO:Plotting -- RooAbsPdf::plotOn(model0) directly selected PDF components: (p_h_sig)
[#1] INFO:Plotting -- RooAbsPdf::plotOn(model0) indirectly selected PDF components: ()
[#1] INFO:Plotting -- RooAbsPdf::plotOn(model0) directly selected PDF components: (p_h_bkg)
[#1] INFO:Plotting -- RooAbsPdf::plotOn(model0) indirectly selected PDF components: ()
[#1] INFO:Plotting -- RooPlot::updateFitRangeNorm: New event count of 50 will supersede previous event count of 1050 for normalization of PDF projections
[#0] WARNING:InputArguments -- RooAbsReal::plotOn(model1) WARNING: argument CurveNameSuffix is duplicated
[#0] WARNING:InputArguments -- RooAbsReal::plotOn(model1) WARNING: argument CurveNameSuffix is duplicated
[#0] WARNING:InputArguments -- RooAbsReal::plotOn(model1) WARNING: argument CurveNameSuffix is duplicated
[#0] WARNING:InputArguments -- RooAbsReal::plotOn(model1) WARNING: argument CurveNameSuffix is duplicated
[#0] WARNING:InputArguments -- RooAbsReal::plotOn(model1) WARNING: argument CurveNameSuffix is duplicated
[#0] WARNING:InputArguments -- RooAbsReal::plotOn(model1) WARNING: argument CurveNameSuffix is duplicated
[#0] WARNING:InputArguments -- RooAbsReal::plotOn(model1) WARNING: argument CurveNameSuffix is duplicated
[#0] WARNING:InputArguments -- RooAbsReal::plotOn(model1) WARNING: argument CurveNameSuffix is duplicated
[#0] WARNING:InputArguments -- RooAbsReal::plotOn(model1) WARNING: argument CurveNameSuffix is duplicated
[#0] WARNING:InputArguments -- RooAbsReal::plotOn(model1) WARNING: argument CurveNameSuffix is duplicated
[#0] WARNING:InputArguments -- RooAbsReal::plotOn(model1) WARNING: argument CurveNameSuffix is duplicated
[#0] WARNING:InputArguments -- RooAbsReal::plotOn(model1) WARNING: argument CurveNameSuffix is duplicated
[#0] WARNING:InputArguments -- RooAbsReal::plotOn(model1) WARNING: argument CurveNameSuffix is duplicated
[#0] WARNING:InputArguments -- RooAbsReal::plotOn(model1) WARNING: argument CurveNameSuffix is duplicated
[#0] WARNING:InputArguments -- RooAbsReal::plotOn(model1) WARNING: argument CurveNameSuffix is duplicated
[#0] WARNING:InputArguments -- RooAbsReal::plotOn(model1) WARNING: argument CurveNameSuffix is duplicated
[#0] WARNING:InputArguments -- RooAbsReal::plotOn(model1) WARNING: argument CurveNameSuffix is duplicated
[#0] WARNING:InputArguments -- RooAbsReal::plotOn(model1) WARNING: argument CurveNameSuffix is duplicated
[#0] WARNING:InputArguments -- RooAbsReal::plotOn(model1) WARNING: argument CurveNameSuffix is duplicated
[#0] WARNING:InputArguments -- RooAbsReal::plotOn(model1) WARNING: argument CurveNameSuffix is duplicated
[#0] WARNING:InputArguments -- RooAbsReal::plotOn(model1) WARNING: argument CurveNameSuffix is duplicated
[#0] WARNING:InputArguments -- RooAbsReal::plotOn(model1) WARNING: argument CurveNameSuffix is duplicated
[#0] WARNING:InputArguments -- RooAbsReal::plotOn(model1) WARNING: argument CurveNameSuffix is duplicated
[#0] WARNING:InputArguments -- RooAbsReal::plotOn(model1) WARNING: argument CurveNameSuffix is duplicated
[#0] WARNING:InputArguments -- RooAbsReal::plotOn(model1) WARNING: argument CurveNameSuffix is duplicated
[#0] WARNING:InputArguments -- RooAbsReal::plotOn(model1) WARNING: argument CurveNameSuffix is duplicated
[#0] WARNING:InputArguments -- RooAbsReal::plotOn(model1) WARNING: argument CurveNameSuffix is duplicated
[#0] WARNING:InputArguments -- RooAbsReal::plotOn(model1) WARNING: argument CurveNameSuffix is duplicated
[#0] WARNING:InputArguments -- RooAbsReal::plotOn(model1) WARNING: argument CurveNameSuffix is duplicated
[#0] WARNING:InputArguments -- RooAbsReal::plotOn(model1) WARNING: argument CurveNameSuffix is duplicated
[#0] WARNING:InputArguments -- RooAbsReal::plotOn(model1) WARNING: argument CurveNameSuffix is duplicated
[#0] WARNING:InputArguments -- RooAbsReal::plotOn(model1) WARNING: argument CurveNameSuffix is duplicated
[#0] WARNING:InputArguments -- RooAbsReal::plotOn(model1) WARNING: argument CurveNameSuffix is duplicated
[#0] WARNING:InputArguments -- RooAbsReal::plotOn(model1) WARNING: argument CurveNameSuffix is duplicated
[#0] WARNING:InputArguments -- RooAbsReal::plotOn(model1) WARNING: argument CurveNameSuffix is duplicated
[#0] WARNING:InputArguments -- RooAbsReal::plotOn(model1) WARNING: argument CurveNameSuffix is duplicated
[#0] WARNING:InputArguments -- RooAbsReal::plotOn(model1) WARNING: argument CurveNameSuffix is duplicated
[#0] WARNING:InputArguments -- RooAbsReal::plotOn(model1) WARNING: argument CurveNameSuffix is duplicated
[#0] WARNING:InputArguments -- RooAbsReal::plotOn(model1) WARNING: argument CurveNameSuffix is duplicated
[#0] WARNING:InputArguments -- RooAbsReal::plotOn(model1) WARNING: argument CurveNameSuffix is duplicated
[#0] WARNING:InputArguments -- RooAbsReal::plotOn(model1) WARNING: argument CurveNameSuffix is duplicated
[#0] WARNING:InputArguments -- RooAbsReal::plotOn(model1) WARNING: argument CurveNameSuffix is duplicated
[#0] WARNING:InputArguments -- RooAbsReal::plotOn(model1) WARNING: argument CurveNameSuffix is duplicated
[#0] WARNING:InputArguments -- RooAbsReal::plotOn(model1) WARNING: argument CurveNameSuffix is duplicated
[#0] WARNING:InputArguments -- RooAbsReal::plotOn(model1) WARNING: argument CurveNameSuffix is duplicated
[#0] WARNING:InputArguments -- RooAbsReal::plotOn(model1) WARNING: argument CurveNameSuffix is duplicated
[#0] WARNING:InputArguments -- RooAbsReal::plotOn(model1) WARNING: argument CurveNameSuffix is duplicated
[#0] WARNING:InputArguments -- RooAbsReal::plotOn(model1) WARNING: argument CurveNameSuffix is duplicated
[#0] WARNING:InputArguments -- RooAbsReal::plotOn(model1) WARNING: argument CurveNameSuffix is duplicated
[#0] WARNING:InputArguments -- RooAbsReal::plotOn(model1) WARNING: argument CurveNameSuffix is duplicated
[#0] WARNING:InputArguments -- RooAbsReal::plotOn(model1) WARNING: argument CurveNameSuffix is duplicated
[#0] WARNING:InputArguments -- RooAbsReal::plotOn(model1) WARNING: argument CurveNameSuffix is duplicated
[#0] WARNING:InputArguments -- RooAbsReal::plotOn(model1) WARNING: argument CurveNameSuffix is duplicated
[#0] WARNING:InputArguments -- RooAbsReal::plotOn(model1) WARNING: argument CurveNameSuffix is duplicated
[#0] WARNING:InputArguments -- RooAbsReal::plotOn(model1) WARNING: argument CurveNameSuffix is duplicated
[#0] WARNING:InputArguments -- RooAbsReal::plotOn(model1) WARNING: argument CurveNameSuffix is duplicated
[#0] WARNING:InputArguments -- RooAbsReal::plotOn(model1) WARNING: argument CurveNameSuffix is duplicated
[#0] WARNING:InputArguments -- RooAbsReal::plotOn(model1) WARNING: argument CurveNameSuffix is duplicated
[#0] WARNING:InputArguments -- RooAbsReal::plotOn(model1) WARNING: argument CurveNameSuffix is duplicated
[#0] WARNING:InputArguments -- RooAbsReal::plotOn(model1) WARNING: argument CurveNameSuffix is duplicated
[#0] WARNING:InputArguments -- RooAbsReal::plotOn(model1) WARNING: argument CurveNameSuffix is duplicated
[#0] WARNING:InputArguments -- RooAbsReal::plotOn(model1) WARNING: argument CurveNameSuffix is duplicated
[#0] WARNING:InputArguments -- RooAbsReal::plotOn(model1) WARNING: argument CurveNameSuffix is duplicated
[#0] WARNING:InputArguments -- RooAbsReal::plotOn(model1) WARNING: argument CurveNameSuffix is duplicated
[#0] WARNING:InputArguments -- RooAbsReal::plotOn(model1) WARNING: argument CurveNameSuffix is duplicated
[#1] INFO:Plotting -- RooAbsPdf::plotOn(model1) directly selected PDF components: (p_ph_sig)
[#1] INFO:Plotting -- RooAbsPdf::plotOn(model1) indirectly selected PDF components: (sp_ph)
[#1] INFO:Plotting -- RooAbsPdf::plotOn(model1) directly selected PDF components: (p_ph_bkg)
[#1] INFO:Plotting -- RooAbsPdf::plotOn(model1) indirectly selected PDF components: (sp_ph)
[#1] INFO:Plotting -- RooPlot::updateFitRangeNorm: New event count of 50 will supersede previous event count of 1050 for normalization of PDF projections
[#0] WARNING:InputArguments -- RooAbsReal::plotOn(model2) WARNING: argument CurveNameSuffix is duplicated
[#0] WARNING:InputArguments -- RooAbsReal::plotOn(model2) WARNING: argument CurveNameSuffix is duplicated
[#0] WARNING:InputArguments -- RooAbsReal::plotOn(model2) WARNING: argument CurveNameSuffix is duplicated
[#0] WARNING:InputArguments -- RooAbsReal::plotOn(model2) WARNING: argument CurveNameSuffix is duplicated
[#0] WARNING:InputArguments -- RooAbsReal::plotOn(model2) WARNING: argument CurveNameSuffix is duplicated
[#0] WARNING:InputArguments -- RooAbsReal::plotOn(model2) WARNING: argument CurveNameSuffix is duplicated
[#0] WARNING:InputArguments -- RooAbsReal::plotOn(model2) WARNING: argument CurveNameSuffix is duplicated
[#0] WARNING:InputArguments -- RooAbsReal::plotOn(model2) WARNING: argument CurveNameSuffix is duplicated
[#0] WARNING:InputArguments -- RooAbsReal::plotOn(model2) WARNING: argument CurveNameSuffix is duplicated
[#0] WARNING:InputArguments -- RooAbsReal::plotOn(model2) WARNING: argument CurveNameSuffix is duplicated
[#0] WARNING:InputArguments -- RooAbsReal::plotOn(model2) WARNING: argument CurveNameSuffix is duplicated
[#0] WARNING:InputArguments -- RooAbsReal::plotOn(model2) WARNING: argument CurveNameSuffix is duplicated
[#0] WARNING:InputArguments -- RooAbsReal::plotOn(model2) WARNING: argument CurveNameSuffix is duplicated
[#0] WARNING:InputArguments -- RooAbsReal::plotOn(model2) WARNING: argument CurveNameSuffix is duplicated
[#0] WARNING:InputArguments -- RooAbsReal::plotOn(model2) WARNING: argument CurveNameSuffix is duplicated
[#0] WARNING:InputArguments -- RooAbsReal::plotOn(model2) WARNING: argument CurveNameSuffix is duplicated
[#0] WARNING:InputArguments -- RooAbsReal::plotOn(model2) WARNING: argument CurveNameSuffix is duplicated
[#0] WARNING:InputArguments -- RooAbsReal::plotOn(model2) WARNING: argument CurveNameSuffix is duplicated
[#0] WARNING:InputArguments -- RooAbsReal::plotOn(model2) WARNING: argument CurveNameSuffix is duplicated
[#0] WARNING:InputArguments -- RooAbsReal::plotOn(model2) WARNING: argument CurveNameSuffix is duplicated
[#0] WARNING:InputArguments -- RooAbsReal::plotOn(model2) WARNING: argument CurveNameSuffix is duplicated
[#0] WARNING:InputArguments -- RooAbsReal::plotOn(model2) WARNING: argument CurveNameSuffix is duplicated
[#0] WARNING:InputArguments -- RooAbsReal::plotOn(model2) WARNING: argument CurveNameSuffix is duplicated
[#0] WARNING:InputArguments -- RooAbsReal::plotOn(model2) WARNING: argument CurveNameSuffix is duplicated
[#0] WARNING:InputArguments -- RooAbsReal::plotOn(model2) WARNING: argument CurveNameSuffix is duplicated
[#0] WARNING:InputArguments -- RooAbsReal::plotOn(model2) WARNING: argument CurveNameSuffix is duplicated
[#0] WARNING:InputArguments -- RooAbsReal::plotOn(model2) WARNING: argument CurveNameSuffix is duplicated
[#0] WARNING:InputArguments -- RooAbsReal::plotOn(model2) WARNING: argument CurveNameSuffix is duplicated
[#0] WARNING:InputArguments -- RooAbsReal::plotOn(model2) WARNING: argument CurveNameSuffix is duplicated
[#0] WARNING:InputArguments -- RooAbsReal::plotOn(model2) WARNING: argument CurveNameSuffix is duplicated
[#0] WARNING:InputArguments -- RooAbsReal::plotOn(model2) WARNING: argument CurveNameSuffix is duplicated
[#0] WARNING:InputArguments -- RooAbsReal::plotOn(model2) WARNING: argument CurveNameSuffix is duplicated
[#0] WARNING:InputArguments -- RooAbsReal::plotOn(model2) WARNING: argument CurveNameSuffix is duplicated
[#0] WARNING:InputArguments -- RooAbsReal::plotOn(model2) WARNING: argument CurveNameSuffix is duplicated
[#0] WARNING:InputArguments -- RooAbsReal::plotOn(model2) WARNING: argument CurveNameSuffix is duplicated
[#0] WARNING:InputArguments -- RooAbsReal::plotOn(model2) WARNING: argument CurveNameSuffix is duplicated
[#0] WARNING:InputArguments -- RooAbsReal::plotOn(model2) WARNING: argument CurveNameSuffix is duplicated
[#0] WARNING:InputArguments -- RooAbsReal::plotOn(model2) WARNING: argument CurveNameSuffix is duplicated
[#0] WARNING:InputArguments -- RooAbsReal::plotOn(model2) WARNING: argument CurveNameSuffix is duplicated
[#0] WARNING:InputArguments -- RooAbsReal::plotOn(model2) WARNING: argument CurveNameSuffix is duplicated
[#0] WARNING:InputArguments -- RooAbsReal::plotOn(model2) WARNING: argument CurveNameSuffix is duplicated
[#0] WARNING:InputArguments -- RooAbsReal::plotOn(model2) WARNING: argument CurveNameSuffix is duplicated
[#0] WARNING:InputArguments -- RooAbsReal::plotOn(model2) WARNING: argument CurveNameSuffix is duplicated
[#0] WARNING:InputArguments -- RooAbsReal::plotOn(model2) WARNING: argument CurveNameSuffix is duplicated
[#0] WARNING:InputArguments -- RooAbsReal::plotOn(model2) WARNING: argument CurveNameSuffix is duplicated
[#0] WARNING:InputArguments -- RooAbsReal::plotOn(model2) WARNING: argument CurveNameSuffix is duplicated
[#0] WARNING:InputArguments -- RooAbsReal::plotOn(model2) WARNING: argument CurveNameSuffix is duplicated
[#0] WARNING:InputArguments -- RooAbsReal::plotOn(model2) WARNING: argument CurveNameSuffix is duplicated
[#0] WARNING:InputArguments -- RooAbsReal::plotOn(model2) WARNING: argument CurveNameSuffix is duplicated
[#0] WARNING:InputArguments -- RooAbsReal::plotOn(model2) WARNING: argument CurveNameSuffix is duplicated
[#0] WARNING:InputArguments -- RooAbsReal::plotOn(model2) WARNING: argument CurveNameSuffix is duplicated
[#0] WARNING:InputArguments -- RooAbsReal::plotOn(model2) WARNING: argument CurveNameSuffix is duplicated
[#0] WARNING:InputArguments -- RooAbsReal::plotOn(model2) WARNING: argument CurveNameSuffix is duplicated
[#0] WARNING:InputArguments -- RooAbsReal::plotOn(model2) WARNING: argument CurveNameSuffix is duplicated
[#0] WARNING:InputArguments -- RooAbsReal::plotOn(model2) WARNING: argument CurveNameSuffix is duplicated
[#1] INFO:Plotting -- RooAbsPdf::plotOn(model2) directly selected PDF components: (p_ph_sig2)
[#1] INFO:Plotting -- RooAbsPdf::plotOn(model2) indirectly selected PDF components: (sp_ph)
[#1] INFO:Plotting -- RooAbsPdf::plotOn(model2) directly selected PDF components: (p_ph_bkg2)
[#1] INFO:Plotting -- RooAbsPdf::plotOn(model2) indirectly selected PDF components: (sp_ph)
[#1] INFO:Plotting -- RooPlot::updateFitRangeNorm: New event count of 50 will supersede previous event count of 1050 for normalization of PDF projections
Asig [normal ] = 0.8337778709310433 +/- 0.1898141852388937
Asig [BB ] = 0.8503293171898778 +/- 0.23578281150991715
Asig [BBlight] = 0.8344736023864808 +/- 0.20295518710344612