

Use Morphing in RooFit.
This tutorial shows how to use template morphing inside RooFit. As input we have several Gaussian distributions. The output is one gaussian, with a specific mean value. Since likelihoods are often used within the framework of morphing, we provide a way to estimate the negative log likelihood (nll).
Based on example of Kyle Cranmer https://gist.github.com/cranmer/46fff8d22015e5a26619.
import ROOT
n_samples = 1000
hist = workspace[f"g{i}"].generateBinned([x_var], n_samples * 100)
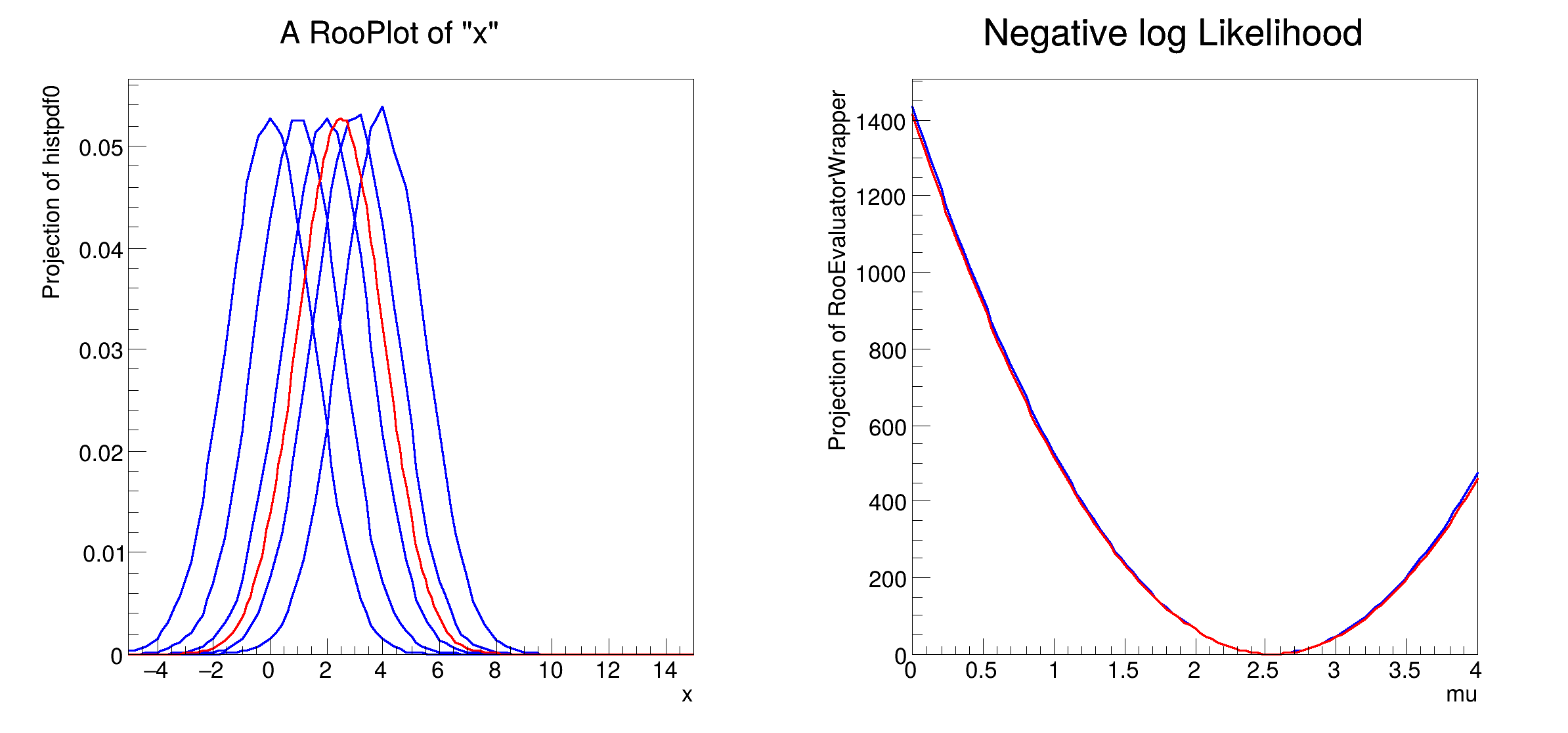
workspace[f"histpdf{i}"].plotOn(frame1)
workspace["morph"].plotOn(frame1, LineColor="r")
return frame1
ws.factory(f
"Gaussian::gauss(x[-5,15], mu[{mu_observed},0,4], {sigma})".
format(mu_observed=mu_observed))
return ws
mu_observed = 2.5
sigma = 1.5
workspace =
build_ws(mu_observed, sigma)
x_var = workspace["x"]
mu_var = workspace["mu"]
nll_morph = workspace["morph"].createNLL(obs_data, EvalBackend="legacy")
c =
ROOT.TCanvas(
"rf616_morphing",
"rf616_morphing", 800, 400)
for nll in [nll_gauss, nll_morph]:
ROOT::Detail::TRangeCast< T, true > TRangeDynCast
TRangeDynCast is an adapter class that allows the typed iteration through a TCollection.
Option_t Option_t TPoint TPoint const char GetTextMagnitude GetFillStyle GetLineColor GetLineWidth GetMarkerStyle GetTextAlign GetTextColor GetTextSize void char Point_t Rectangle_t WindowAttributes_t Float_t Float_t Float_t Int_t Int_t UInt_t UInt_t Rectangle_t Int_t Int_t Window_t TString Int_t GCValues_t GetPrimarySelectionOwner GetDisplay GetScreen GetColormap GetNativeEvent const char const char dpyName wid window const char font_name cursor keysym reg const char only_if_exist regb h Point_t winding char text const char depth char const char Int_t count const char ColorStruct_t color const char Pixmap_t Pixmap_t PictureAttributes_t attr const char char ret_data h unsigned char height h Atom_t Int_t ULong_t ULong_t unsigned char prop_list Atom_t Atom_t Atom_t Time_t format
RooFitResult: minimized FCN value: 1862.97, estimated distance to minimum: 2.32702e-05
covariance matrix quality: Full, accurate covariance matrix
Status : MINIMIZE=0
Floating Parameter FinalValue +/- Error
-------------------- --------------------------
mu 2.5399e+00 +/- 4.74e-02
RooFitResult: minimized FCN value: 1862.82, estimated distance to minimum: 9.31473e-06
covariance matrix quality: Full, accurate covariance matrix
Status : MINIMIZE=0
Floating Parameter FinalValue +/- Error
-------------------- --------------------------
mu 2.5372e+00 +/- 4.08e-02
- Date
- August 2024
- Author
- Robin Syring
Definition in file rf616_morphing.py.


