import ROOT
x = ROOT.RooRealVar("x", "x", -5, 5)
y = ROOT.RooRealVar("y", "y", -5, 5)
z = ROOT.RooRealVar("z", "z", -5, 5)
gx = ROOT.RooGaussian("gx", "gx", x, 0.0, 1.0)
gy = ROOT.RooGaussian("gy", "gy", y, 0.0, 1.0)
gz = ROOT.RooGaussian("gz", "gz", z, 0.0, 1.0)
sig = ROOT.RooProdPdf("sig", "sig", [gx, gy, gz])
px = ROOT.RooPolynomial("px", "px", x, [-0.1, 0.004])
py = ROOT.RooPolynomial("py", "py", y, [0.1, -0.004])
pz = ROOT.RooPolynomial("pz", "pz", z)
bkg = ROOT.RooProdPdf("bkg", "bkg", [px, py, pz])
fsig = ROOT.RooRealVar("fsig", "signal fraction", 0.1, 0.0, 1.0)
model = ROOT.RooAddPdf("model", "model", [sig, bkg], [fsig])
data = model.generate({x, y, z}, 200000)
model.fitTo(data, NumCPU=4, Timer=True, PrintLevel=-1)
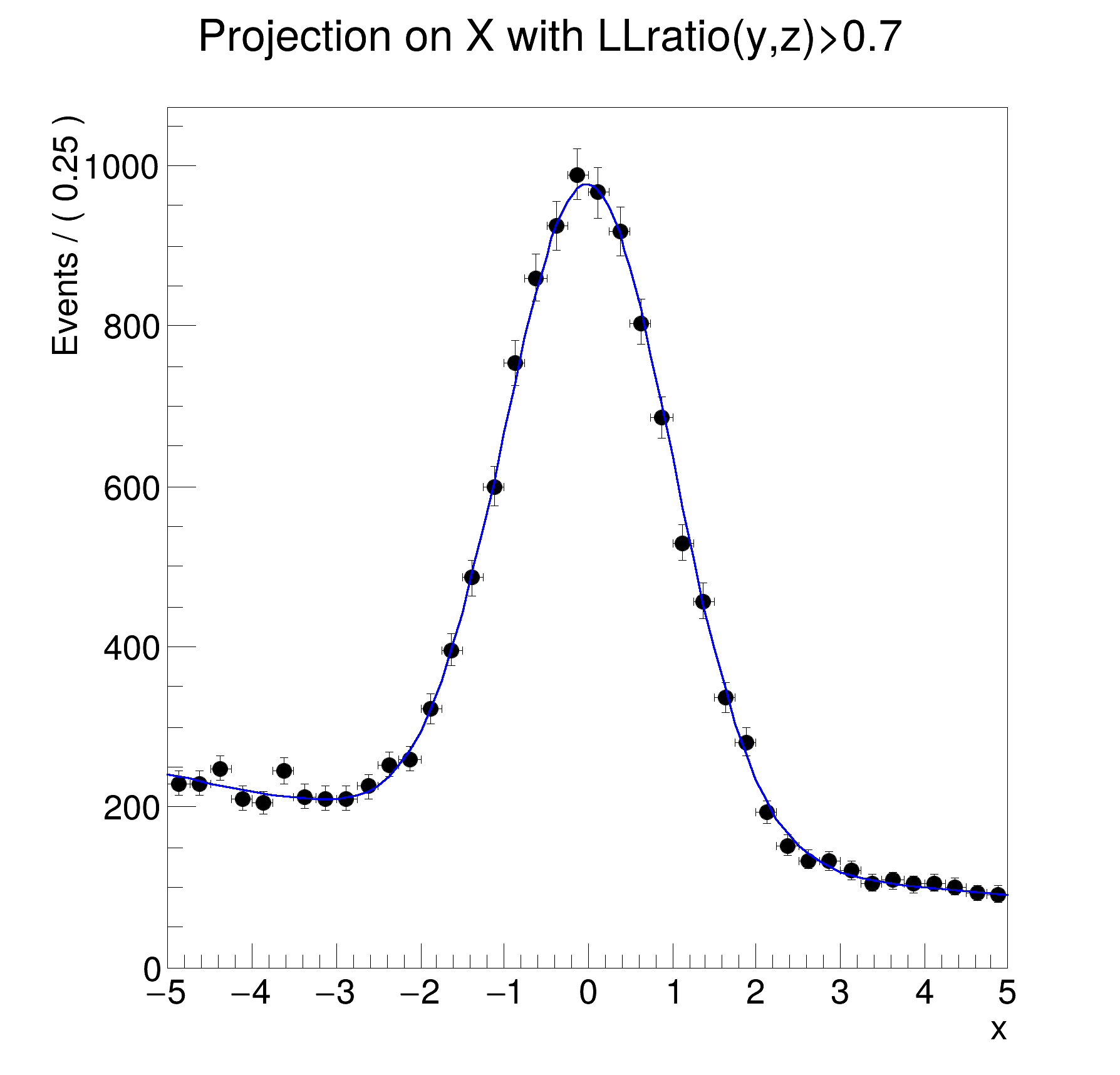
sigyz = sig.createProjection({x})
totyz = model.createProjection({x})
llratio_func = ROOT.RooFormulaVar("llratio", "log10(@0)-log10(@1)", [sigyz, totyz])
data.addColumn(llratio_func)
dataSel = data.reduce(Cut="llratio>0.7")
frame = x.frame(Title="Projection on X with LLratio(y,z)>0.7", Bins=40)
dataSel.plotOn(frame)
model.plotOn(frame, ProjWData=dataSel, NumCPU=4)
c = ROOT.TCanvas("rf603_multicpu", "rf603_multicpu", 600, 600)
ROOT.gPad.SetLeftMargin(0.15)
frame.GetYaxis().SetTitleOffset(1.6)
frame.Draw()
c.SaveAs("rf603_multicpu.png")
[#1] INFO:Fitting -- RooAbsPdf::fitTo(model) fixing normalization set for coefficient determination to observables in data
[#1] INFO:Fitting -- using CPU computation library compiled with -mavx2
[#1] INFO:Fitting -- RooAddition::defaultErrorLevel(nll_model_modelData) Summation contains a RooNLLVar, using its error level
[#1] INFO:Minimization -- RooAbsMinimizerFcn::setOptimizeConst: activating const optimization
[#1] INFO:Minimization -- Command timer: Real time 0:00:00, CP time 0.130
[#1] INFO:Minimization -- Session timer: Real time 0:00:00, CP time 0.130
[#1] INFO:Minimization -- Command timer: Real time 0:00:00, CP time 0.010
[#1] INFO:Minimization -- Session timer: Real time 0:00:00, CP time 0.140, 2 slices
[#1] INFO:Minimization -- RooAbsMinimizerFcn::setOptimizeConst: deactivating const optimization
[#1] INFO:Plotting -- RooAbsReal::plotOn(model) plot on x averages using data variables (y,z)
[#1] INFO:Plotting -- RooAbsReal::plotOn(model) only the following components of the projection data will be used: (y,z)