[#1] INFO:Fitting -- RooAbsPdf::fitTo(gauss_over_gauss_Int[x]) fixing normalization set for coefficient determination to observables in data
[#1] INFO:Fitting -- using generic CPU library compiled with no vectorizations
[#1] INFO:Fitting -- Creation of NLL object took 7.32287 ms
[#1] INFO:Fitting -- RooAddition::defaultErrorLevel(nll_gauss_over_gauss_Int[x]_dh) Summation contains a RooNLLVar, using its error level
[#1] INFO:Minimization -- RooAbsMinimizerFcn::setOptimizeConst: activating const optimization
[#1] INFO:Minimization -- [fitFCN] No discrete parameters, performing continuous minimization only
[#1] INFO:Minimization -- RooAbsMinimizerFcn::setOptimizeConst: deactivating const optimization
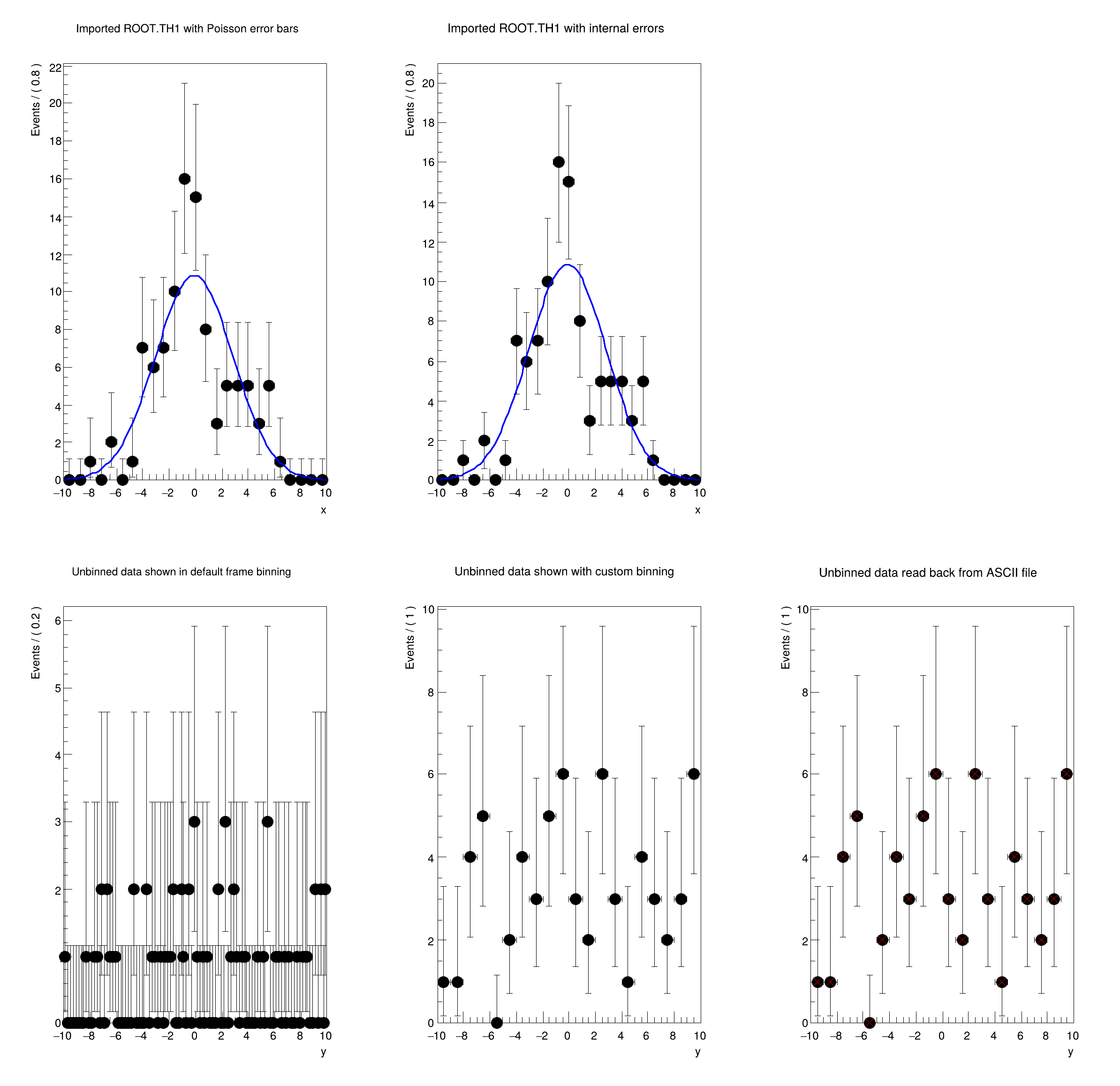
[#1] INFO:DataHandling -- RooTreeDataStore::loadValues(ds) Skipping event #0 because y cannot accommodate the value 14.424
[#1] INFO:DataHandling -- RooTreeDataStore::loadValues(ds) Skipping event #3 because y cannot accommodate the value -12.0022
[#1] INFO:DataHandling -- RooTreeDataStore::loadValues(ds) Skipping event #5 because y cannot accommodate the value 13.8261
[#1] INFO:DataHandling -- RooTreeDataStore::loadValues(ds) Skipping event #6 because y cannot accommodate the value -14.9925
[#1] INFO:DataHandling -- RooTreeDataStore::loadValues(ds) Skipping ...
[#0] WARNING:DataHandling -- RooTreeDataStore::loadValues(ds) Ignored 36 out-of-range events
-----------------------
Reading data from ASCII
[#1] INFO:DataHandling -- RooDataSet::read: reading file rf102_testData.txt
[#1] INFO:DataHandling -- RooDataSet::read: read 64 events (ignored 0 out of range events)
DataStore dataset (rf102_testData.txt)
Contains 64 entries
Observables:
1) x = 9.46654 L(-10 - 10) "x"
2) y = 0.0174204 L(-10 - 10) "y"
3) blindState = Normal(idx = 0)
"Blinding State"
Original data, line 20:
1) RooRealVar:: y = 0.0106407
2) RooRealVar:: x = -0.79919
Read-back data, line 20:
1) RooRealVar:: x = 0.0106407
2) RooRealVar:: y = -0.79919
3) RooCategory:: blindState = Normal(idx = 0)
RooDataSet::ds[y,x] = 64 entries