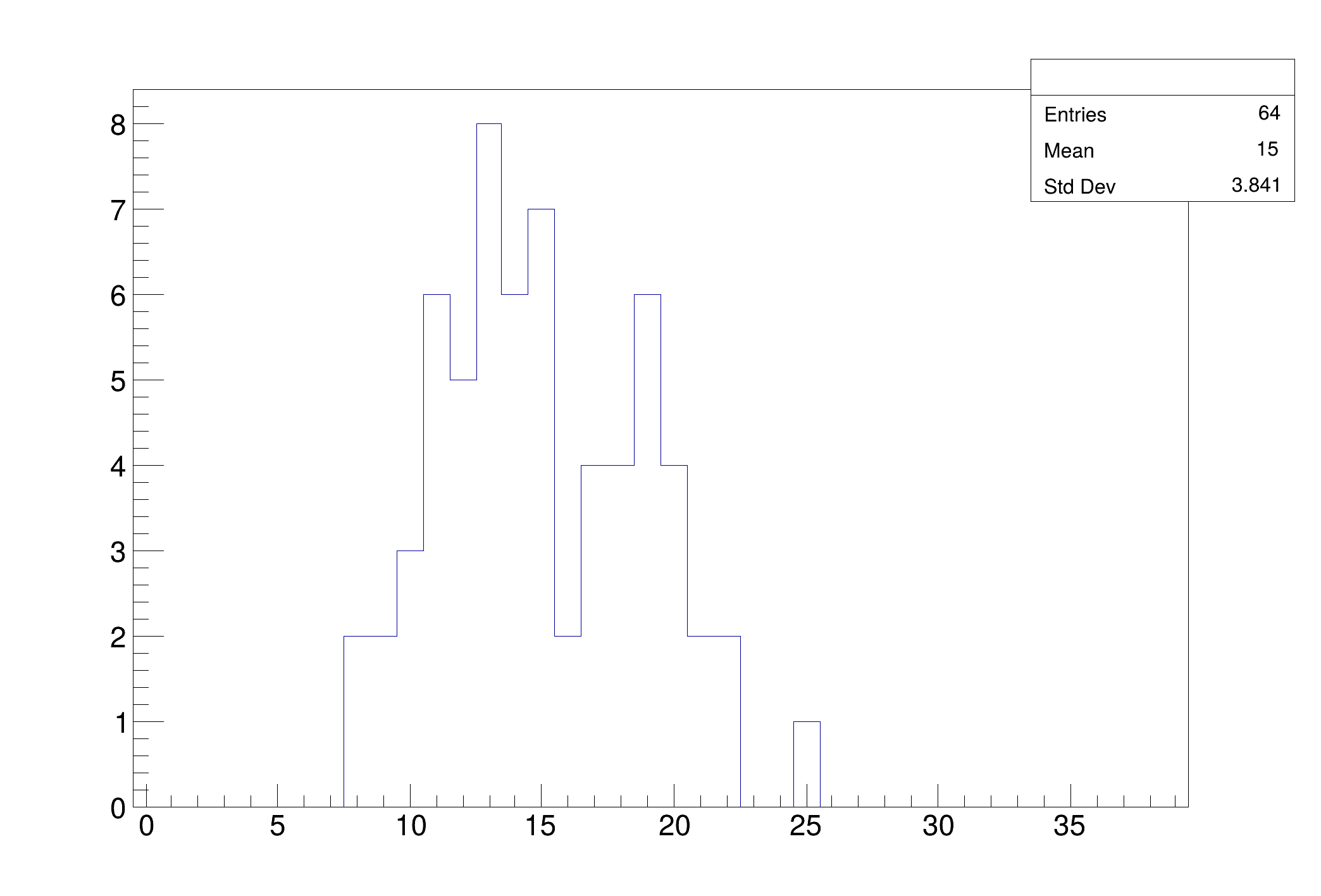
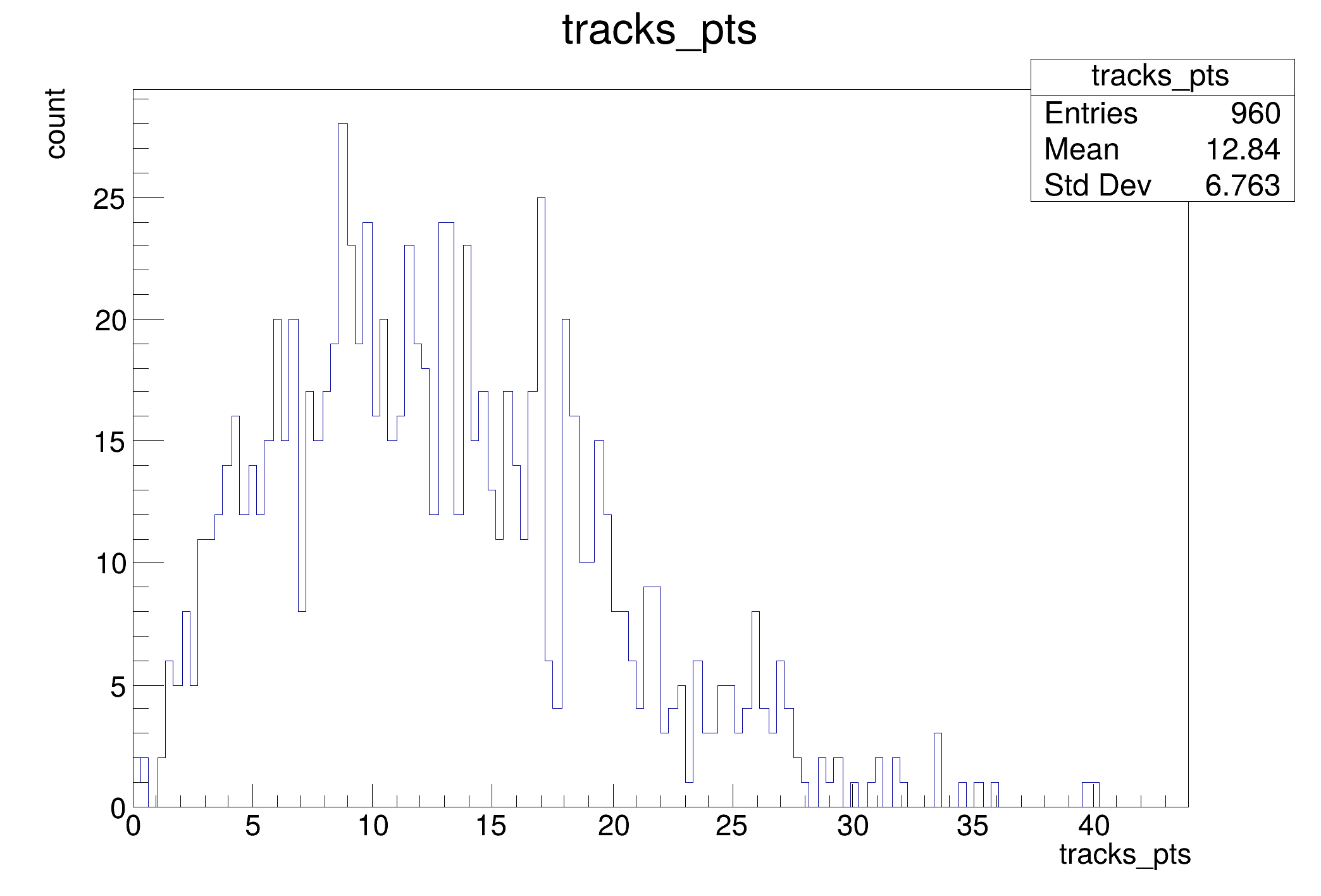
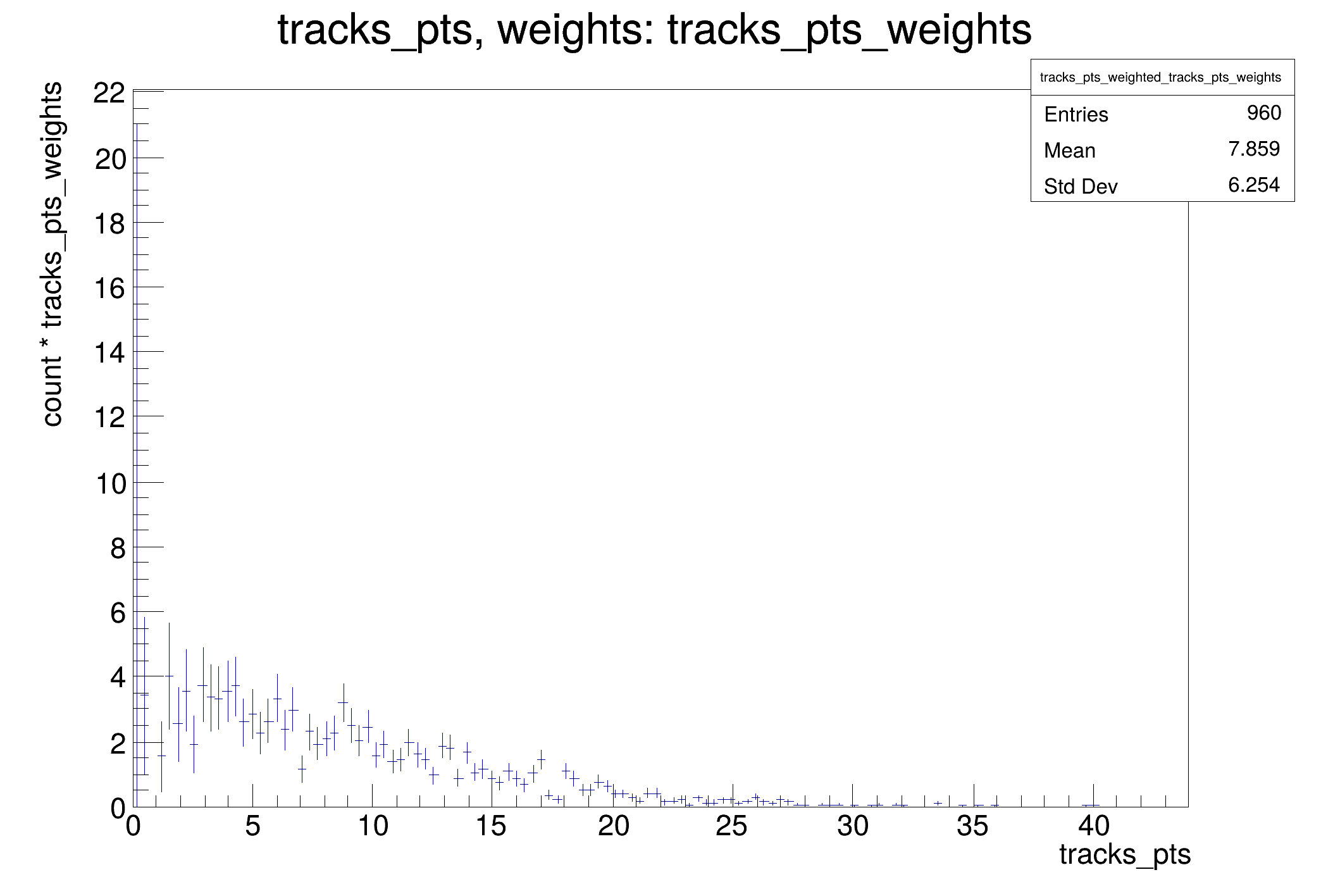
vectors of tracks.
This tutorial shows the possibility to use data models which are more complex than flat ntuples with RDataFrame.
import ROOT
fill_tree_code = """
using FourVector = ROOT::Math::XYZTVector;
using FourVectorVec = std::vector<FourVector>;
using CylFourVector = ROOT::Math::RhoEtaPhiVector;
// A simple helper function to fill a test tree: this makes the example
// stand-alone.
void fill_tree(const char *filename, const char *treeName)
{
const double M = 0.13957; // set pi+ mass
TRandom3 R(1);
auto genTracks = [&](){
FourVectorVec tracks;
const auto nPart = R.Poisson(15);
tracks.reserve(nPart);
for (int j = 0; j < nPart; ++j) {
const auto px = R.Gaus(0, 10);
const auto py = R.Gaus(0, 10);
const auto pt = sqrt(px * px + py * py);
const auto eta = R.Uniform(-3, 3);
const auto phi = R.Uniform(0.0, 2 * TMath::Pi());
CylFourVector vcyl(pt, eta, phi);
// set energy
auto E = sqrt(vcyl.R() * vcyl.R() + M * M);
// fill track vector
tracks.emplace_back(vcyl.X(), vcyl.Y(), vcyl.Z(), E);
}
return tracks;
};
ROOT::RDataFrame d(64);
d.Define("tracks", genTracks).Snapshot(treeName, filename, {"tracks"});
}
"""
fileName = "df002_dataModel_py.root"
treeName = "myTree"
n_cut = "tracks.size() > 8"
getPt_code = """
using namespace ROOT::VecOps;
ROOT::RVecD getPt(const RVec<FourVector> &tracks)
{
auto pt = [](const FourVector &v) { return v.pt(); };
return Map(tracks, pt);
}
"""
getPtWeights_code = """
using namespace ROOT::VecOps;
ROOT::RVecD getPtWeights(const RVec<FourVector> &tracks)
{
auto ptWeight = [](const FourVector &v) { return 1. / v.Pt(); };
return Map(tracks, ptWeight);
};
"""
augmented_d = (
d.Define(
"tracks_n",
"(int)tracks.size()")
.Filter("tracks_n > 2")
.Define("tracks_pts", "getPt( tracks )")
.Define("tracks_pts_weights", "getPtWeights( tracks )")
)
print("Saved figures to df002_*.png")
ROOT::Detail::TRangeCast< T, true > TRangeDynCast
TRangeDynCast is an adapter class that allows the typed iteration through a TCollection.
ROOT's RDataFrame offers a modern, high-level interface for analysis of data stored in TTree ,...