
 Test program as an example for a user specific regularisation scheme
Test program as an example for a user specific regularisation scheme
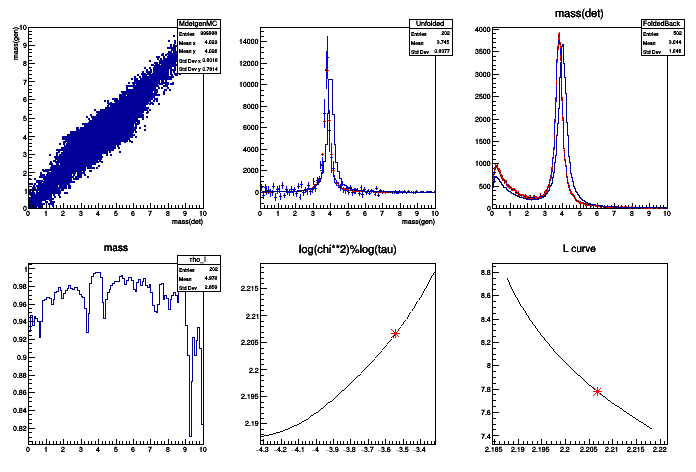
Generate Monte Carlo and Data events The events consist of
The signal is a resonance. It is generated with a Breit-Wigner, smeared by a Gaussian
- Unfold the data. The result is:
- The background level
The shape of the resonance, corrected for detector effects
The regularisation is done on the curvature, excluding the bins near the peak.
- produce some plots
Version 17.0, updated for changed methods in TUnfold
History:
- Version 16.1, parallel to changes in TUnfold
- Version 16.0, parallel to changes in TUnfold
- Version 15, with automatic L-curve scan, simplified example
- Version 14, with changes in TUnfoldSys.cxx
- Version 13, with changes to TUnfold.C
- Version 12, with improvements to TUnfold.cxx
- Version 11, print chi**2 and number of degrees of freedom
- Version 10, with bug-fix in TUnfold.cxx
- Version 9, with bug-fix in TUnfold.cxx, TUnfold.h
- Version 8, with bug-fix in TUnfold.cxx, TUnfold.h
- Version 7, with bug-fix in TUnfold.cxx, TUnfold.h
- Version 6a, fix problem with dynamic array allocation under windows
- Version 6, re-include class MyUnfold in the example
- Version 5, move class MyUnfold to seperate files
- Version 4, with bug-fix in TUnfold.C
- Version 3, with bug-fix in TUnfold.C
- Version 2, with changed ScanLcurve() arguments
- Version 1, remove L curve analysis, use ScanLcurve() method instead
- Version 0, L curve analysis included here
Processing /mnt/vdb/lsf/workspace/root-makedoc-v608/rootspi/rdoc/src/v6-08-00-patches/tutorials/math/testUnfold2.C...
tau=0.000286365
chi**2=160.946+4.9728 / 147
(int) 0
{
do {
do {
} while(t>=1.0);
} while(t<=0.0);
return t;
} else {
do {
do {
} while(t>=1.0);
} while(t>=0.0);
return t;
}
}
return rnd->
Gaus(mTrue+smallBias,smallSigma);
} else {
return rnd->
Gaus(mTrue+wideBias,wideSigma);
}
}
int testUnfold2()
{
TH1D *histMgenMC=
new TH1D(
"MgenMC",
";mass(gen)",nGen,xminGen,xmaxGen);
TH1D *histMdetMC=
new TH1D(
"MdetMC",
";mass(det)",nDet,xminDet,xmaxDet);
TH2D *histMdetGenMC=
new TH2D(
"MdetgenMC",
";mass(det);mass(gen)",nDet,xminDet,xmaxDet,
nGen,xminGen,xmaxGen);
for(
Int_t i=0;i<neventMC;i++) {
4.0,
0.2);
histMgenMC->
Fill(mGen,luminosityData/luminosityMC);
histMdetMC->
Fill(mDet,luminosityData/luminosityMC);
histMdetGenMC->
Fill(mDet,mGen,luminosityData/luminosityMC);
}
TH1D *histMgenData=
new TH1D(
"MgenData",
";mass(gen)",nGen,xminGen,xmaxGen);
TH1D *histMdetData=
new TH1D(
"MdetData",
";mass(det)",nDet,xminDet,xmaxDet);
for(
Int_t i=0;i<neventData;i++) {
3.8,
0.15);
histMgenData->
Fill(mGen);
histMdetData->
Fill(mDet);
}
Int_t iPeek=(
Int_t)(nGen*(estimatedPeakPosition-xminGen)/(xmaxGen-xminGen)
+1.5);
unfold.RegularizeBins(1,1,iPeek-nPeek,regMode);
unfold.RegularizeBins(iPeek+nPeek,1,nGen-(iPeek+nPeek),regMode);
if(unfold.SetInput(histMdetData,0.0)>=10000) {
std::cout<<"Unfolding result may be wrong\n";
}
iBest=unfold.ScanLcurve(nScan,tauMin,tauMax,&lCurve,&logTauX,&logTauY);
std::cout<<"tau="<<unfold.GetTau()<<"\n";
std::cout<<"chi**2="<<unfold.GetChi2A()<<"+"<<unfold.GetChi2L()
<<" / "<<unfold.GetNdf()<<"\n";
for(
Int_t i=1;i<=nGen;i++) binMap[i]=i;
binMap[0]=-1;
binMap[nGen+1]=-1;
TH1D *histMunfold=
new TH1D(
"Unfolded",
";mass(gen)",nGen,xminGen,xmaxGen);
unfold.GetOutput(histMunfold,binMap);
TH1D *histMdetFold=
new TH1D(
"FoldedBack",
"mass(det)",nDet,xminDet,xmaxDet);
unfold.GetFoldedOutput(histMdetFold);
TH1D *histRhoi=
new TH1D(
"rho_I",
"mass",nGen,xminGen,xmaxGen);
unfold.GetRhoI(histRhoi,binMap);
delete[] binMap;
binMap=0;
histMdetGenMC->
Draw(
"BOX");
histMgenData->
Draw(
"SAME");
histMgenMC->
Draw(
"SAME HIST");
histMdetData->
Draw(
"SAME");
histMdetMC->
Draw(
"SAME HIST");
return 0;
}
- Author
- Stefan Schmitt, DESY
Definition in file testUnfold2.C.



 Test program as an example for a user specific regularisation scheme
Test program as an example for a user specific regularisation scheme 