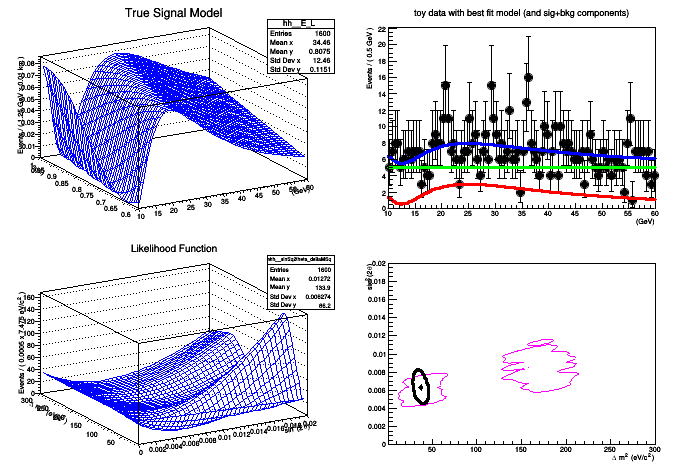
This tutorial shows a more complex example using the FeldmanCousins utility to create a confidence interval for a toy neutrino oscillation experiment. The example attempts to faithfully reproduce the toy example described in Feldman & Cousins' original paper, Phys.Rev.D57:3873-3889,1998.
Processing /mnt/vdb/lsf/workspace/root-makedoc-v608/rootspi/rdoc/src/v6-08-00-patches/tutorials/roostats/rs401d_FeldmanCousins.C...
�[1mRooFit v3.60 -- Developed by Wouter Verkerke and David Kirkby�[0m
Copyright (C) 2000-2013 NIKHEF, University of California & Stanford University
All rights reserved, please read http://roofit.sourceforge.net/license.txt
RooMsgService::setStreamStatus() ERROR: invalid stream ID 2
generate toy data with nEvents = 692
**********
** 1 **SET PRINT 1
**********
**********
** 2 **SET NOGRAD
**********
PARAMETER DEFINITIONS:
NO. NAME VALUE STEP SIZE LIMITS
1 deltaMSq 4.00000e+01 1.95000e+01 1.00000e+00 3.00000e+02
2 sinSq2theta 6.00000e-03 2.00000e-03 0.00000e+00 2.00000e-02
**********
** 3 **SET ERR 0.5
**********
**********
** 4 **SET PRINT 1
**********
**********
** 5 **SET STR 1
**********
NOW USING STRATEGY 1: TRY TO BALANCE SPEED AGAINST RELIABILITY
**********
** 6 **MIGRAD 1000 1
**********
FIRST CALL TO USER FUNCTION AT NEW START POINT, WITH IFLAG=4.
START MIGRAD MINIMIZATION. STRATEGY 1. CONVERGENCE WHEN EDM .LT. 1.00e-03
FCN=-1131.15 FROM MIGRAD STATUS=INITIATE 8 CALLS 9 TOTAL
EDM= unknown STRATEGY= 1 NO ERROR MATRIX
EXT PARAMETER CURRENT GUESS STEP FIRST
NO. NAME VALUE ERROR SIZE DERIVATIVE
1 deltaMSq 4.00000e+01 1.95000e+01 1.99953e-01 1.35503e+01
2 sinSq2theta 6.00000e-03 2.00000e-03 2.21072e-01 -1.80161e+00
ERR DEF= 0.5
MIGRAD MINIMIZATION HAS CONVERGED.
MIGRAD WILL VERIFY CONVERGENCE AND ERROR MATRIX.
COVARIANCE MATRIX CALCULATED SUCCESSFULLY
FCN=-1131.34 FROM MIGRAD STATUS=CONVERGED 32 CALLS 33 TOTAL
EDM=8.53317e-08 STRATEGY= 1 ERROR MATRIX ACCURATE
EXT PARAMETER STEP FIRST
NO. NAME VALUE ERROR SIZE DERIVATIVE
1 deltaMSq 3.75389e+01 4.12974e+00 9.32732e-04 7.25755e-03
2 sinSq2theta 6.29097e-03 8.61732e-04 2.04882e-03 6.82825e-04
ERR DEF= 0.5
EXTERNAL ERROR MATRIX. NDIM= 25 NPAR= 2 ERR DEF=0.5
1.706e+01 -1.140e-03
-1.140e-03 7.447e-07
PARAMETER CORRELATION COEFFICIENTS
NO. GLOBAL 1 2
1 0.31971 1.000 -0.320
2 0.31971 -0.320 1.000
**********
** 7 **SET ERR 0.5
**********
**********
** 8 **SET PRINT 1
**********
**********
** 9 **HESSE 1000
**********
COVARIANCE MATRIX CALCULATED SUCCESSFULLY
FCN=-1131.34 FROM HESSE STATUS=OK 10 CALLS 43 TOTAL
EDM=8.52816e-08 STRATEGY= 1 ERROR MATRIX ACCURATE
EXT PARAMETER INTERNAL INTERNAL
NO. NAME VALUE ERROR STEP SIZE VALUE
1 deltaMSq 3.75389e+01 4.12749e+00 3.73093e-05 -8.56559e-01
2 sinSq2theta 6.29097e-03 8.61259e-04 4.09765e-04 -3.79981e-01
ERR DEF= 0.5
EXTERNAL ERROR MATRIX. NDIM= 25 NPAR= 2 ERR DEF=0.5
1.705e+01 -1.133e-03
-1.133e-03 7.439e-07
PARAMETER CORRELATION COEFFICIENTS
NO. GLOBAL 1 2
1 0.31816 1.000 -0.318
2 0.31816 -0.318 1.000
[#1] INFO:Minization -- p.d.f. provides expected number of events, including extended term in likelihood.
[#1] INFO:NumericIntegration -- RooRealIntegral::init(PnmuTonePrime_Int[EPrime,LPrime]) using numeric integrator RooAdaptiveIntegratorND to calculate Int(LPrime,EPrime)
[#1] INFO:NumericIntegration -- RooRealIntegral::init(PnmuTone_Int[E,L]) using numeric integrator RooAdaptiveIntegratorND to calculate Int(L,E)
[#1] INFO:NumericIntegration -- RooRealIntegral::init(PnmuTone_Int[L]_Norm[E,L]) using numeric integrator RooIntegrator1D to calculate Int(L)
Metropolis-Hastings progress: ....................................................................................................
[#1] INFO:Eval -- Proposal acceptance rate: 3.3%
[#1] INFO:Eval -- Number of steps in chain: 165
[#1] INFO:NumericIntegration -- RooRealIntegral::init(product_Int[deltaMSq,sinSq2theta]_Norm[deltaMSq,sinSq2theta]) using numeric integrator RooAdaptiveIntegratorND to calculate Int(deltaMSq,sinSq2theta)
[#0] WARNING:NumericIntegration -- RooAdaptiveIntegratorND::dtor(product) WARNING: Number of suppressed warningings about integral evaluations where target precision was not reached is 1
[#1] INFO:NumericIntegration -- RooRealIntegral::init(product_Int[deltaMSq,sinSq2theta]_Norm[deltaMSq,sinSq2theta]) using numeric integrator RooAdaptiveIntegratorND to calculate Int(deltaMSq,sinSq2theta)
[#1] INFO:Eval -- cutoff = 0.166573, conf = 0.904333
[#0] WARNING:NumericIntegration -- RooAdaptiveIntegratorND::dtor(product) WARNING: Number of suppressed warningings about integral evaluations where target precision was not reached is 1
[#0] WARNING:NumericIntegration -- RooAdaptiveIntegratorND::dtor(PnmuTone) WARNING: Number of suppressed warningings about integral evaluations where target precision was not reached is 628
[#0] WARNING:NumericIntegration -- RooAdaptiveIntegratorND::dtor(PnmuTonePrime) WARNING: Number of suppressed warningings about integral evaluations where target precision was not reached is 628
Real time 0:04:13, CP time 253.870
MCMC actual confidence level: 0.904333
[#1] INFO:Minization -- RooProfileLL::evaluate(nll_model_modelData_Profile[deltaMSq,sinSq2theta]) Creating instance of MINUIT
[#1] INFO:Minization -- RooProfileLL::evaluate(nll_model_modelData_Profile[deltaMSq,sinSq2theta]) determining minimum likelihood for current configurations w.r.t all observable
[#1] INFO:Minization -- RooProfileLL::evaluate(nll_model_modelData_Profile[deltaMSq,sinSq2theta]) minimum found at (deltaMSq=37.5376, sinSq2theta=0.00629099)
..[#1] INFO:Minization -- LikelihoodInterval - Finding the contour of deltaMSq ( 0 ) and sinSq2theta ( 1 )
Real time 0:05:21, CP time 321.730



 'Neutrino Oscillation Example from Feldman & Cousins'
'Neutrino Oscillation Example from Feldman & Cousins'