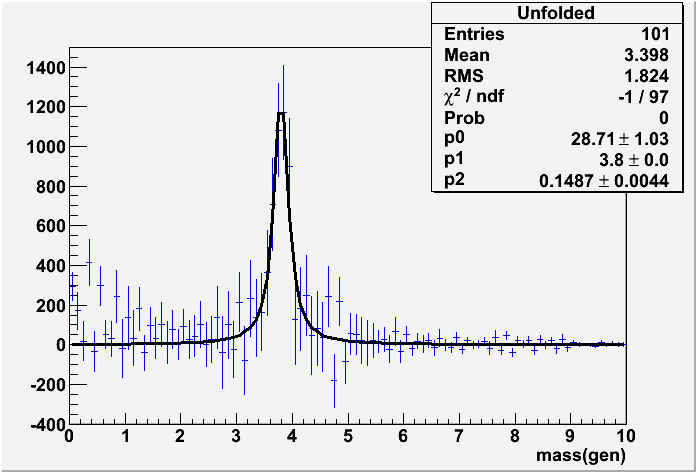
// Test program for the class TUnfold // Author: Stefan Schmitt // DESY, 14.10.2008 // Version 6a, fix problem with dynamic array allocation under windows // // History: // Version 6, bug-fixes in TUnfold.C // Version 5, replace main() by testUnfold1() // Version 4, with bug-fix in TUnfold.C // Version 3, with bug-fix in TUnfold.C // Version 2, with changed ScanLcurve() arguments // Version 1, remove L curve analysis, use ScanLcurve() method instead // Version 0, L curve analysis included here #include <TMath.h> #include <TCanvas.h> #include <TRandom3.h> #include <TFitter.h> #include <TF1.h> #include <TStyle.h> #include <TVector.h> #include <TGraph.h> #include "TUnfold.h" using namespace std; /////////////////////////////////////////////////////////////////////// // // Test program for the class TUnfold // // (1) Generate Monte Carlo and Data events // The events consist of // signal // background // // The signal is a resonance. It is generated with a Breit-Wigner, // smeared by a Gaussian // // (2) Unfold the data. The result is: // The background level // The shape of the resonance, corrected for detector effects // // (3) fit the unfolded distribution, including the correlation matrix // /////////////////////////////////////////////////////////////////////// TRandom *rnd=0; TH2D *gHistInvEMatrix; TVirtualFitter *gFitter=0; void chisquare_corr(Int_t &npar, Double_t * /*gin */, Double_t &f, Double_t *u, Int_t /*flag */) { // Minimization function for H1s using a Chisquare method // only one-dim ensional histograms are supported // Corelated errors are taken from an external inverse covariance matrix // stored in a 2-dimensional histogram Double_t x; TH1 *hfit = (TH1*)gFitter->GetObjectFit(); TF1 *f1 = (TF1*)gFitter->GetUserFunc(); f1->InitArgs(&x,u); npar = f1->GetNpar(); f = 0; Int_t npfit = 0; Int_t nPoints=hfit->GetNbinsX(); Double_t *df=new Double_t[nPoints]; for (Int_t i=0;i<nPoints;i++) { x = hfit->GetBinCenter(i+1); TF1::RejectPoint(kFALSE); df[i] = f1->EvalPar(&x,u)-hfit->GetBinContent(i+1); if (TF1::RejectedPoint()) df[i]=0.0; else npfit++; } for (Int_t i=0;i<nPoints;i++) { for (Int_t j=0;j<nPoints;j++) { f += df[i]*df[j]*gHistInvEMatrix->GetBinContent(i+1,j+1); } } delete[] df; f1->SetNumberFitPoints(npfit); } Double_t bw_func(Double_t *x,Double_t *par) { Double_t dm=x[0]-par[1]; return par[0]/(dm*dm+par[2]*par[2]); } // generate an event // output: // negative mass: background event // positive mass: signal event Double_t GenerateEvent(Double_t const &bgr, // relative fraction of background Double_t const &mass, // peak position Double_t const &gamma) // peak width { Double_t t; if(rnd->Rndm()>bgr) { // generate signal event // with positive mass do { do { t=rnd->Rndm(); } while(t>=1.0); t=TMath::Tan((t-0.5)*TMath::Pi())*gamma+mass; } while(t<=0.0); return t; } else { // generate background event // generate events following a power-law distribution // f(E) = K * TMath::power((E0+E),N0) static Double_t const E0=2.4; static Double_t const N0=2.9; do { do { t=rnd->Rndm(); } while(t>=1.0); // the mass is returned negative // In our example a convenient way to indicate it is a background event. t= -(TMath::Power(1.-t,1./(1.-N0))-1.0)*E0; } while(t>=0.0); return t; } } // smear the event to detector level // input: // mass on generator level (mTrue>0 !) // output: // mass on detector level Double_t DetectorEvent(Double_t const &mTrue) { // smear by double-gaussian static Double_t frac=0.1; static Double_t wideBias=0.03; static Double_t wideSigma=0.5; static Double_t smallBias=0.0; static Double_t smallSigma=0.1; if(rnd->Rndm()>frac) { return rnd->Gaus(mTrue+smallBias,smallSigma); } else { return rnd->Gaus(mTrue+wideBias,wideSigma); } } //int main(int argc, char *argv[]) int testUnfold1() { // switch on histogram errors TH1::SetDefaultSumw2(); // show fit result gStyle->SetOptFit(1111); // random generator rnd=new TRandom3(); // data and MC luminosity, cross-section Double_t const luminosityData=10000; Double_t const luminosityMC=1000000; Double_t const crossSection=1.0; Int_t const nDet=250; Int_t const nGen=100; Double_t const xminDet=0.0; Double_t const xmaxDet=10.0; Double_t const xminGen=0.0; Double_t const xmaxGen=10.0; //============================================ // generate MC distribution // TH1D *histMgenMC=new TH1D("MgenMC",";mass(gen)",nGen,xminGen,xmaxGen); TH1D *histMdetMC=new TH1D("MdetMC",";mass(det)",nDet,xminDet,xmaxDet); TH2D *histMdetGenMC=new TH2D("MdetgenMC",";mass(det);mass(gen)",nDet,xminDet,xmaxDet, nGen,xminGen,xmaxGen); Int_t neventMC=rnd->Poisson(luminosityMC*crossSection); for(Int_t i=0;i<neventMC;i++) { Double_t mGen=GenerateEvent(0.3, // relative fraction of background 4.0, // peak position in MC 0.2); // peak width in MC Double_t mDet=DetectorEvent(TMath::Abs(mGen)); // the generated mass is negative for background // and positive for signal // so it will be filled in the underflow bin // this is very convenient for the unfolding: // the unfolded result will contain the number of background // events in the underflow bin // generated MC distribution (for comparison only) histMgenMC->Fill(mGen,luminosityData/luminosityMC); // reconstructed MC distribution (for comparison only) histMdetMC->Fill(mDet,luminosityData/luminosityMC); // matrix describing how the generator input migrates to the // reconstructed level. Unfolding input. // NOTE on underflow/overflow bins: // (1) the detector level under/overflow bins are used for // normalisation ("efficiency" correction) // in our toy example, these bins are populated from tails // of the initial MC distribution. // (2) the generator level underflow/overflow bins are // unfolded. In this example: // underflow bin: background events reconstructed in the detector // overflow bin: signal events generated at masses > xmaxDet // for the unfolded result these bins will be filled // -> the background normalisation will be contained in the underflow bin histMdetGenMC->Fill(mDet,mGen,luminosityData/luminosityMC); } //============================================ // generate data distribution // TH1D *histMgenData=new TH1D("MgenData",";mass(gen)",nGen,xminGen,xmaxGen); TH1D *histMdetData=new TH1D("MdetData",";mass(det)",nDet,xminDet,xmaxDet); Int_t neventData=rnd->Poisson(luminosityData*crossSection); for(Int_t i=0;i<neventData;i++) { Double_t mGen=GenerateEvent(0.4, // relative fraction of background 3.8, // peak position 0.15); // peak width Double_t mDet=DetectorEvent(TMath::Abs(mGen)); // generated data mass for comparison plots // for real data, we do not have this histogram histMgenData->Fill(mGen); // reconstructed mass, unfolding input histMdetData->Fill(mDet); } //========================================================================= // set up the unfolding // define migration matrix TUnfold unfold(histMdetGenMC,TUnfold::kHistMapOutputVert); // define input and bias scame // do not use the bias, because MC peak may be at the wrong place unfold.SetInput(histMdetData); // the unfolding is done here //=========================== // scan L curve and find best point Int_t nScan=30; Double_t tauMin=1.E-12; Double_t tauMax=1.E-2; Int_t iBest; TSpline *logTauX,*logTauY; TGraph *lCurve; // this method scans the parameter tau and finds the kink in the L curve // finally, the unfolding is done for the best choice of tau iBest=unfold.ScanLcurve(nScan,tauMin,tauMax,&lCurve,&logTauX,&logTauY); std::cout<<"tau="<<unfold.GetTau()<<"\n"; // save graphs with one point to visualize best choice of tau Double_t t[1],x[1],y[1]; logTauX->GetKnot(iBest,t[0],x[0]); logTauY->GetKnot(iBest,t[0],y[0]); TGraph *bestLcurve=new TGraph(1,x,y); TGraph *bestLogTauLogChi2=new TGraph(1,t,x); // set up a bin map, excluding underflow and overflow bins // the bin map maps the output of the unfolding to histogram bins // In this example, the underflow and overflow bin are discarded // This is important for the inverse of the covariance matrix // because that matrix is used for a fit later on Int_t *binMap=new Int_t[nGen+2]; for(Int_t i=1;i<=nGen;i++) binMap[i]=i; binMap[0]=-1; // discarde underflow bin (here: the background normalisation) binMap[nGen+1]=-1; // discarde overflow bin // get unfolded distribution TH1D *histMunfold=new TH1D("Unfolded",";mass(gen)",nGen,xminGen,xmaxGen); unfold.GetOutput(histMunfold,binMap); // get unfolding result, folded back TH1D *histMdetFold=unfold.GetFoldedOutput("FoldedBack","mass(det)", xminDet,xmaxDet); // get matrix of correlation coefficients TH2D *histRhoij=new TH2D("rho_ij",";mass(gen);mass(gen)", nGen,xminGen,xmaxGen,nGen,xminGen,xmaxGen); unfold.GetRhoIJ(histRhoij,binMap); // get global correlation coefficients and inverse of covariance matrix gHistInvEMatrix=new TH2D("invEmat",";mass(gen);mass(gen)", nGen,xminGen,xmaxGen,nGen,xminGen,xmaxGen); TH1D *histRhoi=new TH1D("rho_I","mass",nGen,xminGen,xmaxGen); unfold.GetRhoI(histRhoi,gHistInvEMatrix,binMap); delete[] binMap; binMap=0; // just in case You think it is still defined //====================================================================== // fit Breit-Wigner shape to unfolded data, using the full error matrix // here we use a "user" chi**2 function to take into account // the full covariance matrix //TVirtualFitter::SetDefaultFitter("TMinuit"); gFitter=TVirtualFitter::Fitter(histMunfold); gFitter->SetFCN(chisquare_corr); TF1 *bw=new TF1("bw",bw_func,xminGen,xmaxGen,3); bw->SetParameter(0,1000.); bw->SetParameter(1,3.8); bw->SetParameter(2,0.2); // for (wrong!) fitting without correlations, drop the option "U" histMunfold->Fit(bw,"UE"); //===================================================================== // plot some histograms TCanvas output; // produce some plots output.Divide(3,2); // Show the matrix which connects input and output // There are overflow bins at the bottom, not shown in the plot // These contain the background shape. // The overflow bins to the left and right contain // events which are not reconstructed. These are necessary for proper MC // normalisation output.cd(1); histMdetGenMC->Draw("BOX"); // draw generator-level distribution: // data (red) [for real data this is not available] // MC input (black) [with completely wrong peak position and shape] // unfolded data (blue) output.cd(2); histMunfold->SetLineColor(kBlue); histMunfold->Draw(); histMgenData->SetLineColor(kRed); histMgenData->Draw("SAME"); histMgenMC->Draw("SAME HIST"); // show detector level distributions // data (red) // MC (black) // unfolded data (blue) output.cd(3); histMdetFold->SetLineColor(kBlue); histMdetFold->Draw(); histMdetData->SetLineColor(kRed); histMdetData->Draw("SAME"); histMdetMC->Draw("SAME HIST"); // show correlation coefficients // all bins outside the peak are found to be highly correlated // But they are compatible with zero anyway // If the peak shape is fitted, // these correlations have to be taken into account, see example output.cd(4); histRhoi->Draw("BOX"); // show tau as a function of chi**2 output.cd(5); logTauX->Draw(); bestLogTauLogChi2->SetMarkerColor(kRed); bestLogTauLogChi2->Draw("*"); // show the L curve output.cd(6); lCurve->Draw("AL"); bestLcurve->SetMarkerColor(kRed); bestLcurve->Draw("*"); output.SaveAs("c1.ps"); return 0; }
|