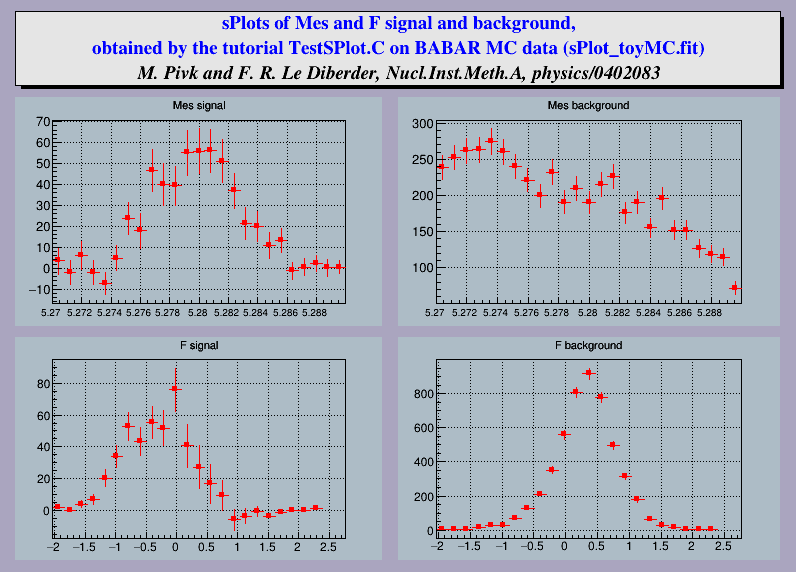
This tutorial illustrates the use of class TSPlot and of the sPlots method.
It is an example of analysis of charmless B decays, performed for BABAR. One is dealing with a data sample in which two species are present: the first is termed signal and the second background. A maximum Likelihood fit is performed to obtain the two yields N1 and N2 The fit relies on two discriminating variables collectively denoted y, which are chosen within three possible variables denoted Mes, dE and F. The variable which is not incorporated in y, is used as the control variable x. The distributions of discriminating variables and more details about the method can be found in the TSPlot class description
NOTE: This script requires a data file $ROOTSYS/tutorials/splot/TestSPlot_toyMC.dat.


Processing /mnt/build/workspace/root-makedoc-v610/rootspi/rdoc/src/v6-10-00-patches/tutorials/splot/TestSPlot.C...
estimated #of events in species 0 = 462.641575
estimated #of events in species 1 = 4957.409256
estimated #of events in species 0 = 490.068958
estimated #of events in species 1 = 4929.942571
estimated #of events in species 0 = 431.484129
estimated #of events in species 1 = 4988.519463
estimated #of events in species 0 = 420.453703
estimated #of events in species 1 = 4999.547303
void TestSPlot()
{
TTree *datatree =
new TTree(
"datatree",
"datatree");
datatree->ReadFile(dataFile,
"Mes/D:dE/D:F/D:MesSignal/D:MesBackground/D:dESignal/D:dEBackground/D:FSignal/D:FBackground/D",' ');
"Mes:dE:F:MesSignal:dESignal:FSignal:MesBackground:"
"dEBackground:FBackground");
ne[0]=500; ne[1]=5000;
"sPlots of Mes and F signal and background", 800, 600);
pt->
AddText(
"sPlots of Mes and F signal and background,");
pt->
AddText(
"obtained by the tutorial TestSPlot.C on BABAR MC " "data (sPlot_toyMC.fit)");
"M. Pivk and F. R. Le Diberder, Nucl.Inst.Meth.A, physics/0402083");
TPad* pad1 =
new TPad(
"pad1",
"Mes signal",0.02,0.43,0.48,0.83,33);
TPad* pad2 =
new TPad(
"pad2",
"Mes background",0.5,0.43,0.98,0.83,33);
TPad* pad3 =
new TPad(
"pad3",
"F signal", 0.02, 0.02, 0.48, 0.41,33);
TPad* pad4 =
new TPad(
"pad4",
"F background", 0.5, 0.02, 0.98, 0.41,33);
}
- Authors
- Anna Kreshuk, Muriel Pivc
Definition in file TestSPlot.C.